Size exclusion chromatography (SEC) is a common technique for characterization of fragments and aggregates in biomolecular samples. Ideally, elution from SEC is governed by hydrodynamic radius rather than molar mass. Traditional SEC depends on reference standards in order to calibrate column elution time as a function of molar mass, assuming that the analytes of interest have the same conformation and chemical composition as the standards.
Nevertheless, the separation of real-world samples often cannot be represented accurately by these standards. Even if the conformation is similar to that of standards (which is rarely the case), real samples may exhibit column interactions that alter their elution properties relative to the standard. When the SEC is coupled with a light scattering detector like the miniDAWN™ TREOS®, it can quantify absolutely the molar mass of a molecule regardless of elution time.
Ultra-High Performance Liquid Chromatography (UHPLC)
UHPLC facilitates rapid, efficient separation of biomolecular samples. UHP-SEC has several advantages over standard SEC, such as higher throughput, improved resolution, less solvent consumption, and smaller sample volume for analysis of valuable biological samples. Detectors intended for standard HPLC cannot be used for UHP-SEC due to excessive dispersion. The Wyatt µDAWNTM multi-angle light scattering (MALS) detector (Figure 1) and Optilab® UT-rEXTM refractive index (RI) detector are specially designed for UHPLC applications. This combination enables measuring the absolute molecular weight (or molar mass) and size of eluting species in UHPLC.

Figure 1. The Wyatt µDAWN MALS Detector.
Advantages of UHPLC Analysis with µDAWN and Optilab UT-rEX
Less than 5 minutes are necessary to complete an entire UHPLC analysis (Figure 2, red) compared to conventional HPLC (Figure 2, blue), which typically takes 30 minutes or more for each sample. In both cases, MALS measures the light scattering intensity and RI the concentration, used in the calculation of molar mass for each peak.
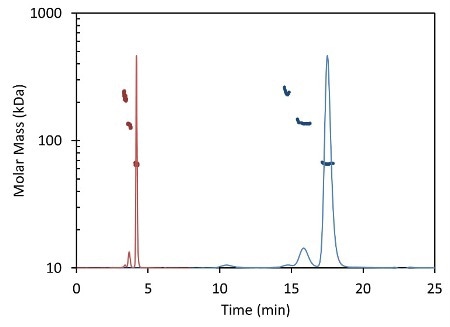
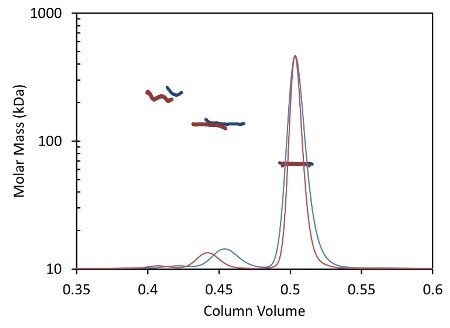
Figure 2. Light scattering data and measured molar mass for a protein separated by UHPLC and detected using the µDAWN and UT-rEX (red) overlaid with the separation by standard HPLC and detected with the miniDAWN TREOS and Optilab T-rEX (blue).
Figure 2 overlays the chromatograms with the monomer, dimer and aggregate molar masses as measured by the miniDAWN TREOS and Optilab T-rEXTM for standard SEC- MALS, and by the µDAWN and Optilab UT-rEX for UHPLC-SEC-MALS. The top plot in Figure 2 delineates the chromatograms as a function of elution time. The same data are rescaled in the bottom plot in Figure 2 as a function of column volume to facilitate comparison. The perfect agreement in molar mass for each peak reveals the possibility of migrating existing HPLC techniques to UHPLC without compromising the accuracy and quality of the molecular weight data.
Besides shortened analysis time, the UHPLC stationary phase with a bead size below 2 µm improves the resolution of aggregate and fragment peaks. Dispersion due to the large internal volume of standard columns and detectors broadens out these tiny peaks, causing the ‘vanishing’ of some poorly-resolved species like the fragment illustrated in Figure 3 as blue traces.
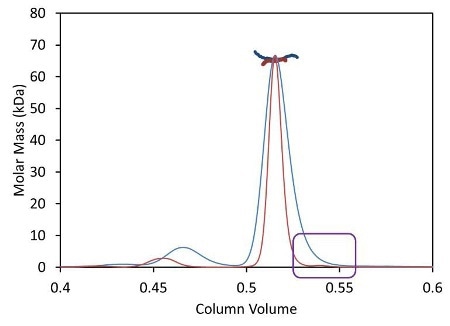
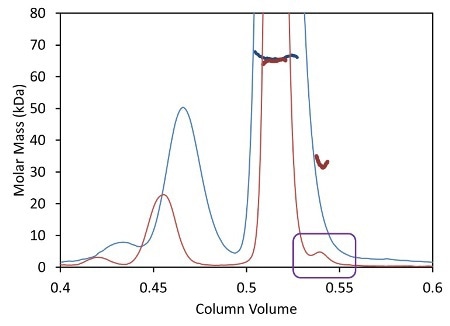
Figure 3. Light scattering data and measured molar mass for a protein separated by UHPLC (red) overlaid with the separation by standard HPLC (blue).
The µDAWN is engineered for least internal volume to maintain the resolution in a UHPLC elution profile, thereby facilitating the detection and investigation of previously unknown species. The box in Figure 3 shows a fragment peak that can be separated only by UHPLC. The bottom plot in Figure 3 shows a zoomed-in view revealing the fragment and its molar mass, as measured by the µDAWN.
Fragment analysis is greatly enhanced by RI detection, a universally applicable means of online concentration measurement which depends only on the refractive index increment (dn/dc) of the analyte relative to the solvent. The refractive index increment is virtually constant for all proteins. Hence, unlike UV-based concentration measurements, it is not necessary to determine the identity of the fragment in order to ascertain the extinction coefficient of the peak in order to quantify the eluting concentration and molar mass. Actually, the combination of UV and RI detection may be employed to know the extinction coefficient of each species and help in their identification.
Conclusion
The µDAWN not only measures absolute molecular weight, but also provides all the in-depth analyses performed with the miniDAWN TREOS to UHPLC. As with standard HPLC-MALS, variations in molar mass across the UHPLC-MALS peak measure polydispersity of a biopolymer or validate reversible protein oligomerization. Triple detection with UV, MALS, and RI facilitates Protein Conjugate Analysis to determine the degree of post-translational modification, related detergent, drug-conjugate, or other additions to a poly-peptide background.
The µDAWN replicates the 10-50 nm range of rms radius (Rg, radius of gyration) available to the three-angle miniDAWN. In addition, optional dynamic light scattering (DLS) detection with the WyattQELS™ module quantifies hydrodynamic radius from 0.5 to 40 nm in the same measurement volume as MALS detection.
Reference
Adapted from "Absolute Molar Mass in UHPLC via the µDAWN and UT-rEX," white paper by Wyatt Technology. Graphs and illustrations reprinted with permission from Wyatt Technology.

This information has been sourced, reviewed and adapted from materials provided by Wyatt Technology.
For more information on this source, please visit Wyatt Technology.