Size-exclusion chromatography (SEC) is a chromatographic technique which is often utilized in the biological science field.
This method is usually adopted to determine the molecular weight of a sample by referencing the elution time of the unknown molecular weight versus the standards of the known molecular weight. This procedure is performed using a concentration detector such as refractive index (RI) or ultraviolet (UV) .
Now, sophisticated multi-detector SEC solutions come with up to four detectors such as RI, UV, intrinsic viscosity (LV) and light scattering (LS). In this article, a bacterial membrane protein and a purified membrane protein sample are characterized by means of the Viscotek TDAmax system (Figure 1), a comprehensive SEC system with UV, RI, LS and IV detectors.

Figure 1. The TDAmax system.
Light Scattering and Intrinsic Viscosity
Intrinsic viscosity is defined as a measure of molecular density and helps in evaluating structural changes.
A light scattering detector helps in determining the molecular weights and the necessity for column calibration. Intrinsic viscosity together with light scattering enables the measurement of size (Rh). It is important to maintain the detectors so that a high level of sensitivity is maintained across all four detectors.
In molecular biology research, membrane protein studies are increasingly being performed. One of the key objectives of these studies is crystallization of proteins. However, this process has proved to be very complex, because crystallization of membrane proteins relies on a number of factors such as purity of protein and the detergent type or concentration. When excess amount of the detergent component of the protein complex is removed, it can degrade the protein and thus reduce the possibility of crystallization.
Therefore, both the characterization and optimization of the amount of detergent and protein in a purified sample of membrane protein can provide a better understanding about the possibility of crystallization and also about the content of protein in the protein detergent complex (PDC).
Experimental Framework
Sample purification is done using the n-dodecyl ß-D-maltoside (DDM) detergent. The PBS added with 0.02% w/v DDM served as the mobile phase and the sample was isolated on a GE Superdex 200 SEC column.
The OmniSEC co-polymer analysis was utilized which employs the dA/dc and dn/dc of the detergent and the protein to determine the concentration and molecular weight of both the components at each one of the data slice along the chromatogram. For the dn/dc of the protein, a value of 0.185ml/g was utilized and its extinction coefficient was predicted to be 0.5 (A280; 1mg/ml). Prior study has demonstrated the dn/dc of DDM to be of 0.1608ml/g value and its extinction coefficient was calculated to be 0.0044 (A280; 1mg/ml).
Results and Discussion
Given the fact that only a small quantity of purified protein is available for examination, bovine serum albumin (BSA) was used to study the micelle behavior of protein and detergent on the system. The protein’s molecular weight was utilized to calibrate the instrument’s detector responses. Then, a surplus amount of pure DDM micelles was run to determine their size and molecular weight. Chromatograms of DDM, BSA and the two together are shown in Figure 2.
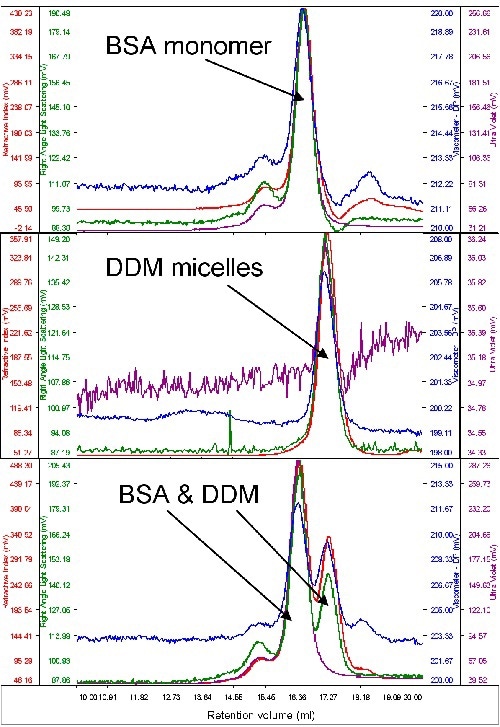
Figure 2. From top to bottom, chromatograms of BSA, DDM and a mixture of the two.
From the above graph, both samples are seen to elute at different volumes, rendering rational resolution between the two. Also, the signal from the DDM is extremely low, whilst BSA displays considerable absorption on the UV channel (280nm). This is obvious from the chromatograms when both the samples were run together and run alone.
The BSA’s molecular weight was calculated to be 67kDa. When measured alone, the DDM micelles had a size of 2.5nm and a quantified molecular weight of 60kDa. When the DDM and BSA were measured simultaneously, a small increase in DDM molecular weight (65kDa) was seen. This may be due to the restricted resolution between the two peaks.
The membrane protein was infused after the DDM and BSA were found to run suitably on the system. Protein concentration was 0.5mg/ml and 100µl was loaded. In one peak, the sample eluted with a shoulder on the ascending edge in accordance with the RI. This shoulder was related to most of the UV signal, distinguishing it as the protein detergent complex (PDC), whilst the peak’s main body had small amount of UV signal, implying that this was the DDM micelles, as shown in Figure 3.
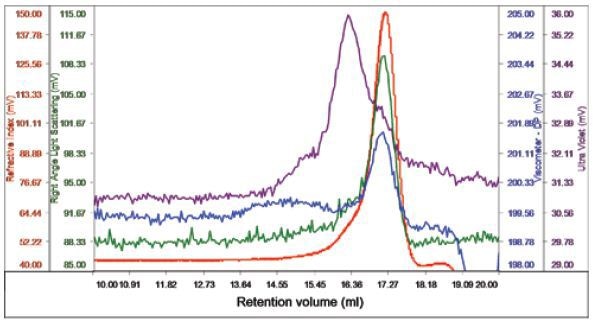
Figure 3. Chromatogram of bacterial multidrug membrane protein.
The peak’s region sans UV absorbance had a size of 2.6nm and a measured molecular weight of 62.5kDa, thus distinguishing it as the DDM micelles. The UV peak and shoulder was recognized as the PDC and had 74.5kDa molecular weight. Upon examining the conjugate, the amount of PDC in the overall sample was measured to be 21.6%, while the DDM micelles contained 78.4% of the specimen. The PDC was quantified to comprise 54% and 46% of DDM and protein, respectively. The protein’s molecular weight was found to be 33kDa, which is near to the estimation that protein contained 46% of the PDC and gives excellent validation for the results obtained.
For each of the two components, the concentration can be measured and plotted in a derived chromatogram displaying concentration of the component in place of detector responses (Figure 4).
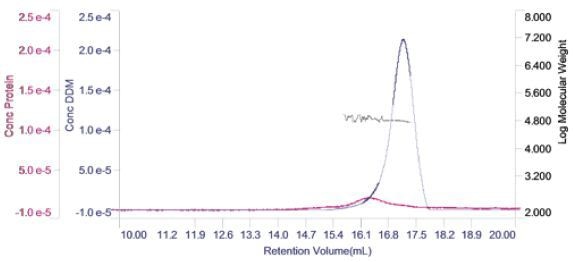
Figure 4. Derived chromatogram showing the concentration of protein (purple) and DDM (blue). Measured molecular weight is plotted across the two peaks.
This gives a good visual representation of the sample’s composition. In this case, it is apparent that the protein’s concentration is considerably lower when compared to the DDM micelles. In addition, it is clear that majority of the DDM is not in the PDC, but in free micelles. Also, on the derived chromatogram, the molecular weight of both the peak has been plotted.
Conclusion
Using the Viscotek TDAmax system, the protein detergent complex was characterized which occurred at the time of the purification of bacterial multi-drug membrane protein in the presence of DDM. The molecular weight of both the free DDM micelles and the PDC was determined. Moreover, the PDC’s composition with respect to DDM and protein content was determined and observed to be close to expectations.

This information has been sourced, reviewed and adapted from materials provided by Malvern Panalytical.
For more information on this source, please visit Malvern Panalytical.