The TopSpin™ software package from Bruker is designed for NMR data analysis and the acquisition and processing of NMR spectra. This software package, TS3.5pl6 is a latest release from Bruker and features a new, easy-to-use GUI, which offers easy access to vast experiment libraries including typical Bruker pulse sequences, and user generated experiment libraries for both academic and industrial users.
The GUI enables users to construct and organize their own tailored experiment libraries via simple drag and drop, enables script based parameter modification, offers a number of options for experiment setup and optimization, and makes the setup of advanced experiments simple and efficient.
TopSpin provides a completely workflow-oriented user interface and exploits the modern 64-bit features of modern Windows / MacOS/ CentOS operating systems for optimum memory usage. Its new workflow-oriented and intuitive user interface and optimized sample data management both provide a long list of extra innovative features, meant exclusively to accelerate operation and sample analysis throughput for better cost efficiency. TopSpin is also provided in a student version if required for helping institutions in education and academia to improve the curricula of their students.
TopSpin was built with a highly intuitive interface applying the most extensive standards familiar from word processing, presentation programs, or graphics, providing the same look-and-feel for NMR applications on Windows®, Mac, and Linux®.
Features
The key features of the TopSpin™ software package are as follows:
- Total control of Avance and Fourier spectrometers
- Comprehensive functionalities for processing, displaying and analyzing multidimensional and single spectra
- Intuitive industry standard user interface, the ideal solution for both regular users and experts
- Unlimited offline processing capabilities
- Assisted acquisition
- Advanced non-uniform sampling techniques
- Small molecule characterization
- BioToolsTM - Biomolecular NMR made simple
- Method development environment
- Simple and efficient structure elucidation of tiny molecules with CMC-seTM
- Lineshape analysis for solid-state NMR, including dynamic NMR
- Result publishing, predefined and user-defined layouts
- Regulatory compliance support tools (audit trailing, auto-archiving, electronic signature)
- Special licenses for universities and students
Technical Details
User Interface
- Complete functionalities for processing, displaying and analyzing multidimensional and single spectra
- Intuitive industry standard user interface, the ideal solution for both standard users and experts
- Assisted acquisition
- Deconvolution/spectrum simulation/iteration
- Result publishing, predefined and user-defined layouts
- Individual user customization (experiments, toolbars, commands, workflows)
- Complete documentation
- Regulatory compliance (GMP, GLP) support tools (electronic signature, audit trailing, auto-archiving)
- PC (Linux / Windows) or Mac
- Flexible licensing for a variety of applications
- Special licenses for universities students and
Supported Operating Systems
TopSpin 3.5 on spectrometer computers is supported for:
- Windows 7 (64 bit)
- CentOS 7.1 (64 bit)
- CentOS 5 (64 bit)
TopSpin 3.5 on data stations is supported for:
- Windows Vista (32-bit)
- Windows 8.1 (32- and 64-bit)
- Windows 7 (32- and 64-bit)
- CentOS 7.1 (64-bit)
- CentOS 5 (32- and 64-bit)
- Mac-OS X 10.8 or newer
To effectively install and operate TopSpin under CentOS, extra packages will be required:
- TopSpin Installation Requirements for CentOS 7
- TopSpin Installation Requirements for CentOS 5
Acquisition and Processing Packages
- BioTools™ - protein NMR made easy
- Solids lineshape analysis
- Non-uniform sampling (option)
- Small molecule structure elucidation (option)
- Structure chemical shift prediction (option)
- Structure confidence analysis (option)
Applications
Absolute Quantification - CMC-q
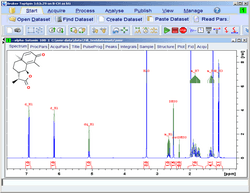
It is possible to perform absolute quantification of a series of samples automatically, thus enabling users to define mass content, purity and relative quantities of substances. This provides a totally automated concentration determination in drug discovery screening applications.
Structure Elucidation CMC-se
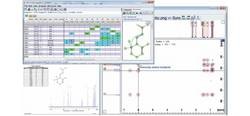
Beginning from a chemical formula and standard NMR experiments, peaks are automatically chosen and entered into a correlation table. This data is then interpreted using a structure-determination algorithm and structures that are consistent with the data are produced. These structures can then be ranked in relation to comparison with chemical shift predictions, thus narrowing down the possibilities, to assist the researcher in establishing their structure.
Non Uniform Sampling
TopSpin 3.0 currently has Non Uniform Sampling (NUS), which is a fully integrated acquisition and processing feature. It allows repetitive use and is available for all kinds of NMR experiments. An automatically generated and optimized NUS sparse list establishes the sampling pattern resulting in the fractional data acquisition. Multi-dimensional decomposition then allows calculation of the missing data points, enabling a regular Fourier Transform processing of the complete data set, all performed in a completely automatic fashion.
NUS eliminates a time barrier for high-resolution multidimensional NMR spectroscopy, and is particularly beneficial for multi-dimensional experiments in biomolecular NMR, where time savings up to a factor of 4 in 3D experiments or 10 and even more in 4D and 5D experiments can be realized. New protein structure determination experiments are now feasible with NUS.
Small molecule applications based on 2D spectra can also gain from faster acquisition by a factor of 2. A key highlight here is that improvements in spectral resolution and quality delivered by NUS are performed without having to increase the overall acquisition time.
NUS implementation in TopSpin results from a research partnership with Prof. Orekhov and co-workers. NUS in TopSpin 3.0 benefits from well-established algorithms and programs such as NUS-Sampler and MDDNMR (Orekhov, V.Y., I. Ibraghimov, and M. Billeter, J. Biomol. NMR, 2003. 27(2):p. 165-173)
Dynamics Center
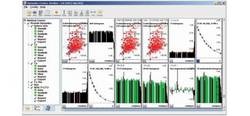
The new Dynamics Center is a technique-oriented workflow that exploits efficient protein data evaluation, providing all applicable dynamic parameters, such as Rex, T1, T2 with minimum user interaction. The new capability is designed mainly for exploring the dynamics of large bio-molecules.