May 9 2016
First successful protein structural analysis in a levitating drop of liquid.
Knowledge of the exact structure of proteins – those biological molecules that perform multifaceted and essential functions in the organism – is crucial, for example, in producing new active ingredients for drugs.
This protein structure is normally deciphered by means of X-radiation through so-called X-ray structural analysis. Soichiro Tsujino and Takashi Tomizaki, two scientists at the Paul Scherrer Institute PSI, have used a clever trick to advance this method further: They have successfully determined the structure of a protein inside a levitating drop of liquid.
They used ultrasound to make the drop hover in the air. With this trick they were able to carry out the structural analysis at room temperature and thus very close to the natural conditions in the organism – a major advantage over the classical method, in which the protein sample is fixed to a specimen holder and also must be cooled to low temperatures. Soichiro Tsujino and Takashi Tomizaki have now published their study in the journal Scientific Reports.
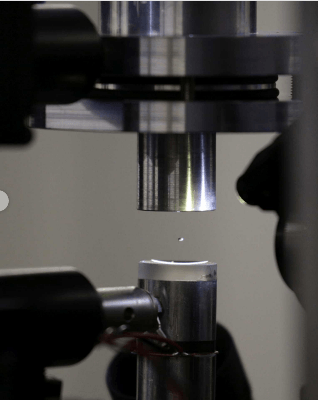 With an ultrasound plate, the researchers caused a tiny drop of fluid, around one millimetre in diameter, to levitate. Freely moving within it: the protein crystal to be analysed. (Photo: Paul Scherrer Institute/Takashi Tomizaki)
With an ultrasound plate, the researchers caused a tiny drop of fluid, around one millimetre in diameter, to levitate. Freely moving within it: the protein crystal to be analysed. (Photo: Paul Scherrer Institute/Takashi Tomizaki)
Two scientists at the Paul Scherrer Institute PSI have chosen an unusual medium for the study of a protein: a levitating drop of liquid. They used ultrasound to make it hover in the air while they simultaneously determined the protein structure with X-rays from the Swiss Light Source SLS. Since the structure of lysozyme, the protein they used, is already known from previous measurements, Soichiro Tsujino and Takashi Tomizaki were able to show that their method leads to the correct structure. With that, the X-ray structural analysis of a protein in a levitating droplet was successfully demonstrated for the first time. The major advantage of the new method is that it can be carried out at room temperature.
Proteins: Structure is crucial
Proteins are found in the cells of every living thing. All in all, countless types of proteins perform diverse functions in the organism. The specific function of a protein is strongly associated with its structure, that is, how the building blocks of a protein are aligned with each other. Accurate knowledge of the protein structure is thus crucial for pursuits such as the development of custom-tailored medicinal agents.
X-ray structural analysis itself, also known as X-ray crystallography, is an established method. For this purpose, first micrometre-sized crystals are grown from numerous copies of a protein molecule.
These crystals are cooled to low temperatures, around minus 170 degrees Celsius, and then probed with a very fine and very intense X-ray beam. Only a few facilities in the world – among them the Swiss Light Source SLS at the PSI – deliver such a beam. X-rays deflected in the sample contain information about the arrangement of atoms and thus about the structure of the particular protein.
In order to evaluate this information, the X-radiation is recorded by a detector and ultimately analysed by computer. At the SLS, three of the twenty-plus measuring stations are specialised for X-ray structural analysis. Here, over the past ten years, researchers were able to decipher the structures of around 4000 different proteins. With that, the SLS ranks as one of the most productive facilities of this kind worldwide.
Sought and found: a room-temperature research method
"The problem with the conventional method is the required cooling of the protein crystals," explains Tomizaki, a biologist. This is done so that the proteins in the crystal will not be damaged by the X-rays.
On the other hand, Tomizaki says, "no one can guarantee that at minus 170 degrees Celsius the proteins still retain their complete natural structure – that is, the structure that they have at body temperature in the organism." That's why he and physicist Soichiro Tsujino were looking for an alternative method that would work at room temperature, and thus very close to the conditions in the living organism.
The two researchers came up with the idea to bring a small drop of liquid containing a protein crystal into the X-ray beam. The researchers dripped a droplet less than a millimetre in diameter – four microlitres of liquid – in the air above a special plate, which they vibrated with ultrasound to make the drop hover in the air. At SLS’s macromolecular crystallography beamline, the researchers were able to analyse the structure of a protein in the droplet at room temperature.
With this acoustic levitation method, they have simultaneously resolved a second difficulty, namely the manipulation of the tiny protein crystals. Classical structural analysis requires the painstaking effort of attaching them to a specimen holder. Tsujino and Tomizaki had it easier: The growth of protein crystals is carried out in a protein solution. With a pipette, the two scientists simply took up a drop of this solution together with the crystal it contained.
"The simplicity of our method makes it, at the same time, a very economical procedure. External scientists who come to the SLS to probe the structure of proteins will appreciate both advantages," Tsujino predicts.
Extremely fast detectors make it possible
The idea of studying a sample suspended in an acoustically levitating drop of liquid is not entirely new. Until now, however, no research team has succeeded in carrying out an effective X-ray structural analysis of a protein in this way.
"We too were only successful in this undertaking because our beamline at the SLS is equipped with detectors with extremely fast data processing," Tsujino says. This capability is needed because the crystal suspended in the droplet does not maintain a fixed orientation, but rather moves constantly and uncontrollably while rotating on its own axis.
In classical X-ray structural analysis, one first obtains a picture of individual bright spots. The structure of the protein can be worked out from the location of these points. "Through the continuous rotation of the crystal in the droplet, however, these points would really be smeared to lines – then classical structural analysis is out of the question," Tsujino explains.
Here the ultrafast detectors from the company DECTRIS – a spin-off of the PSI – delivered the solution. With the EIGER X 16M detector, the researchers were able to capture 133 images per second. The individual exposures were so short that the crystal appeared as if frozen. This can be compared to fast-action sports photography: Here too short exposure times are called for, so that the athlete in the picture won't look blurry.
Uncontrolled crystal motion as a double benefit
It was also no problem for the researchers that the crystal's motion inside the droplet was completely uncontrollable: The X-ray light bent – that is, diffracted – by the crystal contains information about the angle from which the crystal was illuminated. So to the contrary, the rotation of the crystal proved advantageous: After several thousand exposures, the crystal had been illuminated from all sides, so that a perfect structural analysis was possible. "Thousands of pictures may sound like a lot, but thanks to the fast image processing of our detectors, we only needed about half a minute for this," Tsujino says. "Next we want to repeat the experiment with another detector, the EIGER 1M, which can process 3000 images per second. With that we could record all the needed data within around one second."
Moreover, it was the constant rotation and movement of the crystal in the droplet that made it possible to carry out the measurement at room temperature. At10 micrometres, the cross-section of the X-ray beam is considerably smaller than the protein crystals, which at around 200 micrometres are roughly twice the thickness of a blond human hair. Thus the X-ray beam is always striking only a small part of the crystal. The crystal's continuous motion guarantees that no area is exposed to the radiation long enough to be damaged.
"Certainly we could also build a specimen holder that rapidly rotates and moves the crystal. But why bother, when the crystal in the levitating droplet does it all on its own, and much more inexpensively?" says Tsujino, laughing.
Proof of success with a known protein
Tsujino and Tomizaki were able to demonstrate the capability of their new method with the protein lysozyme: Its structure is already known, thanks to classical X-ray structural analysis. In comparison, the researchers could judge that the structure they detected in their levitating droplet was correct.
The researchers were also able to show that in their experiment the proteins in fact did not suffer any radiation damage.
Collaboration with a PSI spin-off
Meanwhile, Tsujino and Tomizaki are also collaborating with another PSI spin-off company: leadXpro AG, based in PARK innovARRE on the PSI campus. Members of the leadXpro staff were fascinated that the PSI researchers' method significantly shortened the time needed for X-ray structural analysis of a protein. Together, the PSI researchers and scientists from leadXpro now want to further develop the new method. This concrete collaboration is currently funded by the Swiss Commission for Technology and Innovation CTI.
Text: Paul Scherrer Institute/Laura Hennemann