Nov 4 2020
Some molecules tend to bind tightly to the surface of the ice, forming a curved interface that inhibits further growth of the ice crystal.
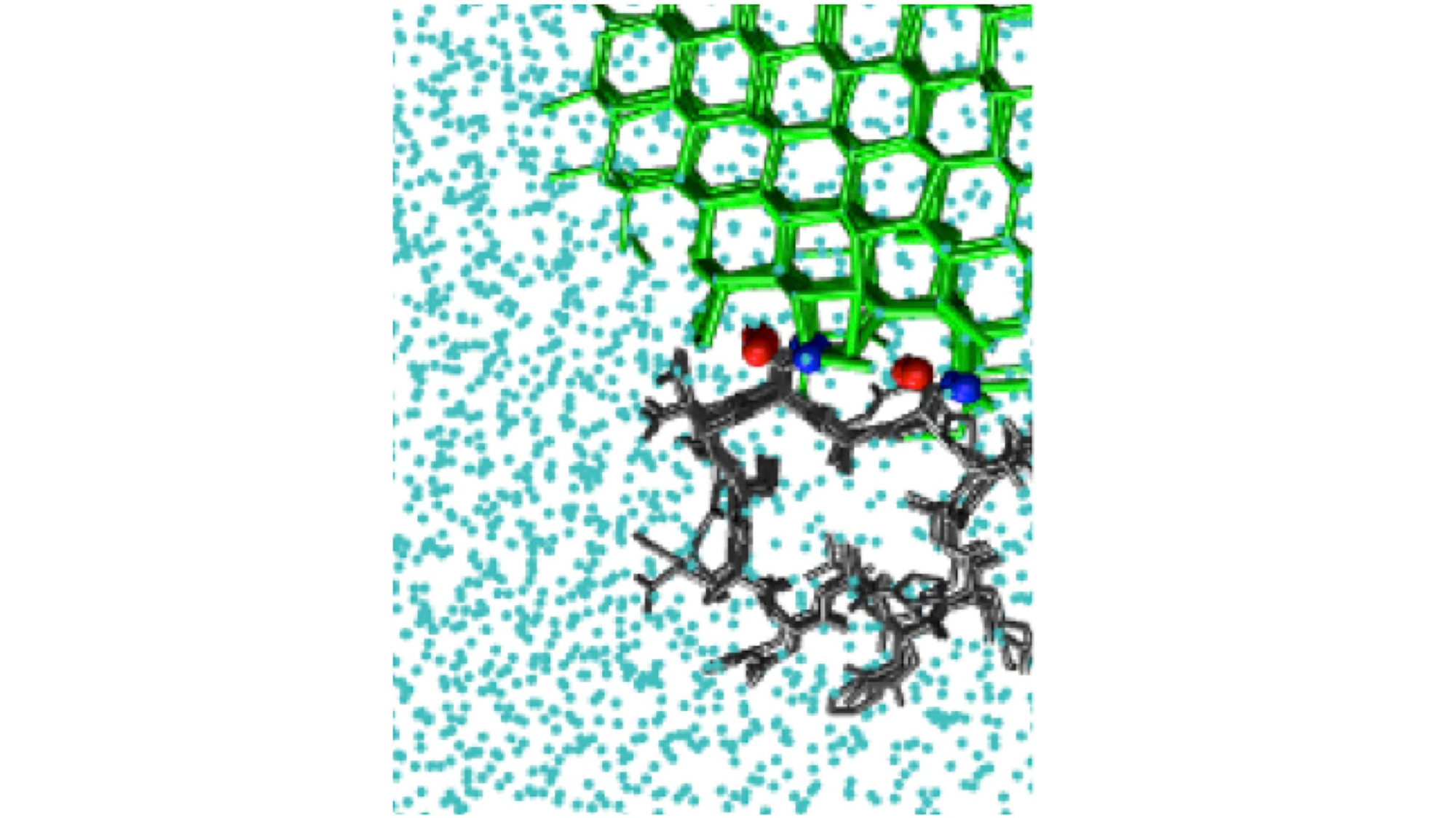 Ice-biasing simulations can detect the ice-binding site of the hyperactive antifreeze protein from the beetle Tenebrio molitor, TmAFP. Image Credit: Pavithra M. Naullage.
Ice-biasing simulations can detect the ice-binding site of the hyperactive antifreeze protein from the beetle Tenebrio molitor, TmAFP. Image Credit: Pavithra M. Naullage.
Certain plants, insects, and aquatic creatures include protein molecules of this kind that serve as natural antifreeze agents, which enable the organisms to endure freezing temperatures.
In the Journal of Chemical Physics, from AIP Publishing, a team of researchers has described a computational technique to model ice-binding, which involves using a biasing method to induce ice formation in the simulation.
Antifreeze proteins function by binding to an interface that exists between liquid water and ice. The ensuing curved surface prevents ice growth. Moreover, there are ice-nucleating molecules that catalyze the development of ice from supercooled liquid water.
Both phenomena need insights into the way molecules bind to ice. Gaining knowledge about ice-binding is essential for applications ranging from climate modeling to cryopreservation of organs. However, at present, there are no computational techniques to efficiently model this phenomenon.
The central advantage of the ice-biasing simulation approach is that it simultaneously identifies the ice-binding surface, the face of ice it binds to, and the mode of binding.
Valeria Molinero, Study Author, University of Utah
Two types of models were made by the investigators. One is an all-atom model that includes all the atoms present in the ice and liquid phases of water, as well as in the antifreeze-type molecule.
The second model studied is known as a coarse-grained model, which reduces the computational resources used by mixing atoms into simpler structures.
The study analyzed several molecules that bind ice, such as polyvinyl alcohol—a synthetic ice-recrystallization inhibitor—and natural antifreeze proteins, like the one from the beetle Tenebrio molitor.
As proteins feature very small surfaces that bind ice, they are difficult to simulate. This restricts the size of the ice crystal that can be bound by them.
Certain systems exhibit more than one location for ice to bind. This holds true for the natural antifreeze protein present in the sea-ice diatom Frailariopsis cylindrus. The researchers designed a technique called “cap and repeat” to identify whether a protein like this has more than one ice-binding surface (IBS).
In this strategy, we first performed a biased simulation to detect an IBS. Then, we cap that IBS to prevent ice formation on it and perform a second biasing simulation to find out whether ice forms in other sites.
Valeria Molinero, Study Author, University of Utah
The techniques developed as part of the study exhibit huge potential for several applications, such as determining molecules to safeguard frozen tissues at the time of storage, gaining further insights into natural antifreeze proteins, as well as in climate models, where ice nucleation in the air plays a vital role.
Journal Reference:
Naullage, P. M., et al. (2020) Computationally efficient approach for the identification of ice-binding surfaces and how they bind ice. The Journal of Chemical Physics. doi.org/10.1063/5.0021631.