Determining alcohols using NMR spectroscopy is carried out in order to introduce students to NMR spectroscopy at an early stage in their undergraduate career. The experiment aims to enable students to detect an alcohol using 1D 13C-NMR and DEPT experiments and allocate the peaks, reiterating previous knowledge of electronegativity and naming of organic compounds.
This experiment enables students to obtain their own spectra for solving the structure of a considerably simple compound with the chance to include distillation for identifying the boiling point of an unknown alcohol as a further experimental technique.
Introduction
In comparison with 1H-NMR spectra, 1H-decoupled 13C-NMR spectra are considerably easier to interpret. There are three key concepts that students are made familiar with early in their undergraduate studies that are needed for the interpretation of 13C-NMR spectra: hybridization, electronegativity and the number of distinct carbons in the structure.
These concepts, introduced in various areas of undergraduate chemistry, can be integrated to understand the number of resonances and chemical shift that are seen in 13C-NMR spectra.
This experiment uses alcohol identification for introducing NMR spectroscopy. Alcohol usage enables students to correspond electronegativity and chemical shift and the 13C-NMR spectrum peaks are suitably separated for direct interpretation.
In order that students use the nomenclature for the naming of organic compounds, alcohols are a simple example for students. Distillation can also be used as an experimental method for purifying and determining the boiling point of organic liquids. All the alcohol samples are liquids, hence, NMR analysis samples can be conveniently prepared as students can obtain spectra of neat liquids.
Using the Spinsolve Carbon system, students can conduct Distortionless Enhancement by Polarisation Transfer (DEPT) experiments and also conventional 13C-NMR spectra. The carbon nuclei can be differentiated based on the number of protons attached to it. All carbons are seen in the 13C-NMR spectra, using DEPT-90 only CH's (3° carbons) and with DEPT-135, protonated carbons are observed, with CH and CH3' positive and CH2's (2° carbons) negative. Each student can determine the alcohol assigned to them from a class list based on the 13C and DEPT spectra. In case, this experiment is the first time the student is exposed to NMR spectroscopy, further boiling point information offers confidence to students with their assignments.
Experiment
Students are given a class list that includes the possible unknown alcohols. In the lab, each student is allocated one unknown alcohol. Distillation is used to ascertain the alcohol’s boiling point that also helps purify the liquid (Figure 1).
Direct sampling of a 0.5 mL aliquot of the purified alcohol is performed into a 5mm NMR tube. The students use the Spinsolve Carbon system to acquire 13C and DEPT data for their alcohol, and then process the data for determining their unknown alcohol and assigning the peaks in the spectrum.
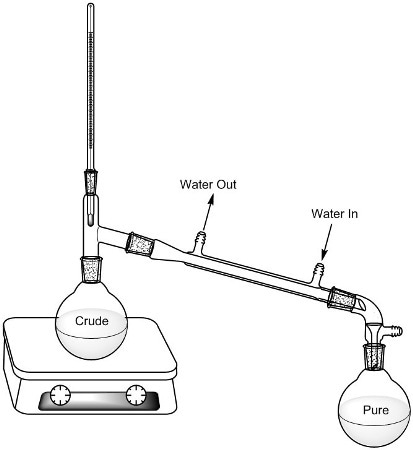
Figure 1. Distillation set-up
Safety
Almost all the unknown alcohols used in the experiment may be poisonous and hazardous when inhaled, ingested, or exposed to skin. Most of the alcohols are flammable, hence the unknowns must not be near flames and also while heating at the time of distillation, it is important to take utmost care. All safety precautions must be taken with any unknown compound.
Potential List of Unknowns
All the alcohols given in Table 1 have a separate set of DEPT spectra, and hence, there must be no ambiguity while assigning. Most of the compounds though, have an identical number of resonances in the 13C-NMR spectrum, showing how DEPT is highly powerful for structural elucidation of compounds.
Table 1. Alcohol list
| Compound |
Structure |
# Carbons |
13C |
CH |
CH2 |
CH3 |
Boiling Point °C |
| Ethanol |
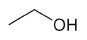 |
2 |
2 |
0 |
1 |
1 |
78 |
| 2-propanol |
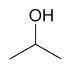 |
3 |
2 |
1 |
0 |
1 |
82 |
| 1-propanol |
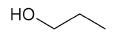 |
3 |
3 |
0 |
2 |
1 |
97 |
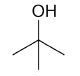
| 2-methyl-2-propanol |
 |
4 |
2 |
0 |
0 |
1 |
82 |
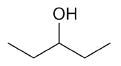
| 3-pentanol |
 |
5 |
3 |
1 |
1 |
1 |
115 |
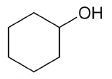
| cyclohexanol |
 |
6 |
4 |
1 |
3 |
0 |
160 |
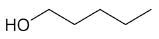
| 1-pentanol |
 |
5 |
5 |
0 |
4 |
1 |
138 |
| 1-hexanol |
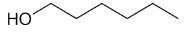 |
6 |
6 |
0 |
5 |
1 |
157 |
| 2-methylcyclohexanol |
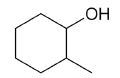 |
7 |
7 |
2 |
4 |
1 |
165 |
| 2,4-dimethyl-3-pentanol |
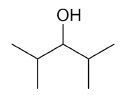 |
7 |
3 |
2 |
0 |
1 |
140 |
| cyclopentanol |
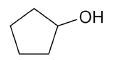 |
5 |
3 |
1 |
2 |
0 |
140 |
| 2-methyl-2-butanol |
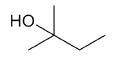 |
5 |
4 |
0 |
1 |
2 |
102 |
| 1-butanol |
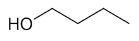 |
4 |
4 |
0 |
3 |
1 |
117 |
| 3,3-dimethyl-2-butanol |
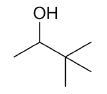 |
6 |
4 |
1 |
0 |
2 |
120 |
| 4-methyl-2-pentanol |
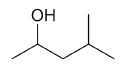 |
6 |
5 |
2 |
1 |
2 |
132 |
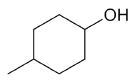
| 4-methylcyclohexanol |
 |
7 |
5 |
2 |
2 |
1 |
170 |
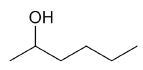
| 2-hexanol |
 |
6 |
6 |
1 |
3 |
2 |
140 |
Tasks and Questions
- Firstly purify, and then ascertain the unknown alcohol’s boiling point by means of distillation.
- Record the 1D 13C-NMR, DEPT 135 and DEPT 90 spectra of the unknown alcohol.
- Determine the unknown alcohol using information collated, and mention the reasons for assigning that structure to the unknown alcohol.
- Assign the corresponding spectrum peaks to carbon environments in the alcohol, elucidating the chemical shifts of the carbon environments.
Example Spectra
Figure 2 shows the spectra acquired for 1-butanol, which was performed in two experiments, and stacked during data processing.
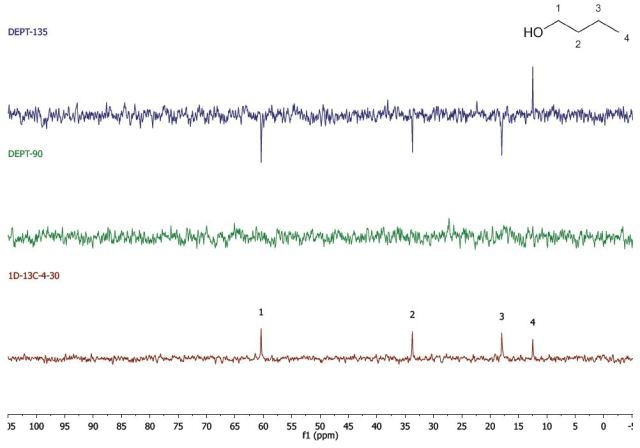
Figure 2. DEPT and 1D 13C-NMR spectra of neat 1-butanol (4 scans)
The 1D 13C-NMR experiment was carried out by means of four scans with a 30s repetition time. Hence, the spectrum was obtained in below 2 mins. In the same way, the spectrum in the DEPT experiment was acquired with 4 scans and a 30 s repetition time. Since there are two spectra obtained, the experiment was completed in 4 mins. Hence, the total acquisition time for the spectra in Figure 2 was 6 mins.
The Spinsolve software is convenient and quick. Hence, there is a need for minimal additional time while setting up the experiment. The experiment needs to be chosen with a series of buttons: input the sample name and choose the parameters (Figure 3). It is also possible to automate the experiment sequence using Scripts, so that students only need to press the start button. Sample preparation is also very rapid and easy, hence, the total time spent by the students with the instrument may be around 10 mins.
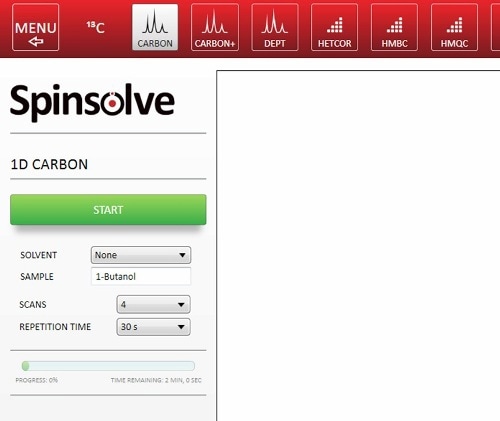
Figure 3. Setting up a 1D 13C-NMR spectrum in the Spinsolve software.
In case there is a need for a higher throughput, the 1D 13C-NMR spectra can be obtained in a single scan. The acquisition time is brought down to 30 s. Further, the repetition time may be 10 s for the DEPT experiments, bringing down the total acquisition time to around 1.5 min (Figure 4).
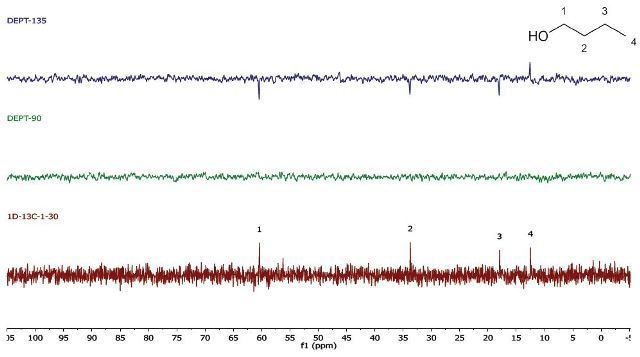
Figure 4. EPT (4 scans, 10s repetition time) and 1D 13C-NMR (1 scan, 30 s repetition time) spectra of neat 1-butanol
Note that for the 1D 13C-NMR spectrum, a repetition time of 30s is needed because of the long T1 relaxation time of almost all carbon environments. The DEPT experiment depends on polarization transfer from 1H to 13C. Hence, its repetition time is controlled by the much shorter T1 of protons. If need be, it is also possible to bring down the carbon T1 by introducing a relaxation agent, which enables much faster repetition times.
While using whichever spectra set, it can be observed that there are four carbon environments (1D- 13C), there are no tertiary carbons (DEPT-90), and there are three CH2's and one CH3 (DEPT-135). Students use this data along with the principles of electronegativity to allocate the chemical shifts in the molecule they have detected.
Propan-2-ol
DEPT and 1D 13C-NMR spectra for Propan-2-ol are shown in Figure 5.
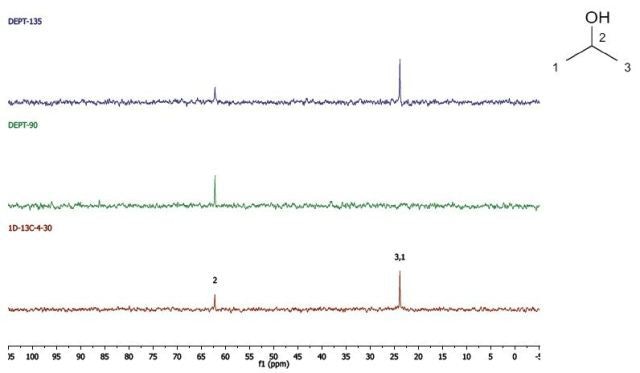
Figure 5. DEPT and 1D 13C-NMR spectra of neat propan-2-ol (4 scans).
Cyclohexanol
DEPT and 1D 13C-NMR spectra of neat cyclohexanol are shown in Figure 6.
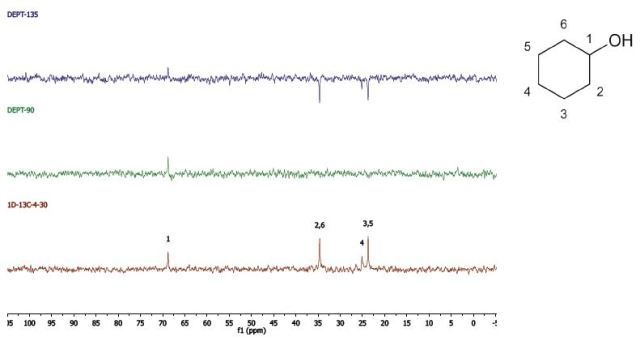
Figure 6. DEPT and 1D 13C-NMR spectra of neat cyclohexanol (4 scans)
Since, cyclohexanol and Propan-2-ol have an axis of symmetry about the hydroxyl group they hence show lesser resonances in the 13C-NMR spectrum than there are carbons in the molecule. Students can thus think of the concept of a range of chemical environments recognizing that symmetry causes several nuclei to have the same chemical shift.
2-Methyl-2-propanol
DEPT and 1D 13C-NMR spectra of neat 2-methyl-2-propanol are shown in Figure 7.
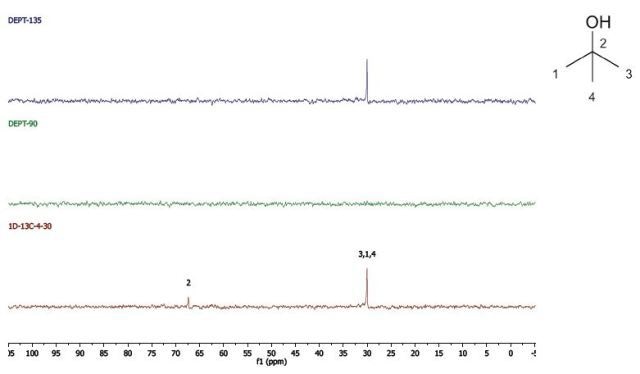
Figure 7. DEPT and 1D 13C-NMR spectra of neat 2-methyl-2-propanol (4 scans)
2-Methyl-2-propanol is an instance of a molecule having a quartnernary carbon. This kind of carbon is seen in the 13C-NMR spectrum, however, as it is not protonated, it is not seen in the DEPT spectra.
Ethanol
Figure 8 shows the DEPT and 1D 13C-NMR spectra of neat ethanol.
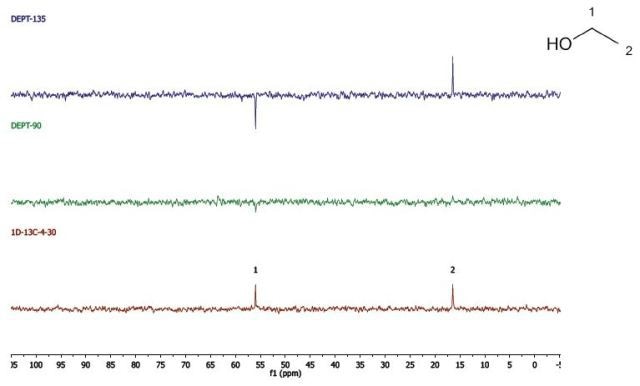
Figure 8. DEPT and 1D 13C-NMR spectra of neat ethanol (4 scans).
This information has been sourced, reviewed and adapted from materials provided by Magritek.
For more information on this source, please visit Magritek.