Limited by the laws of physics, scientific discoveries and the advance of knowledge can be bound up in a series of slow proceedings. In microscopy, up until the early 21st century, the diffraction limit of light was a seemingly impenetrable barrier. It was believed, in line with the law of physics, that two points could not be resolved (separated) if the distance between them is less than half the wavelength of light used to observe them.
Effectively, this determines that in practice light microscopes could only resolve objects/structures that have a separating distance of 200 nm or more. Therefore, since many subcellular structures and organelles are smaller than 200 nm, the 200 nm barrier would present a considerable gap of knowledge waiting to be uncovered.
During the first decade of the 21st century, scientists and researchers successfully solved the obstacle presented by the diffraction limit of light using a series of innovative microscopy techniques: a new field of discovery was emerging. Imaging beyond the diffraction limit of light evolved in parallel with the development of new methodologies for super-resolution.
Super-resolution microscopy methods that included; STORM/PALM (Stochastic optical reconstruction microscopy)/ (Fluorescence photo-activation localization microscopy); STED (stimulated emission depletion microscopy); and SIM (Structured illumination microscopy) are techniques readily available and accessible to researchers.
While the super-resolution methods have opened a new field of discovery, some restrictions on these methods were presented. High energy requirements for acquisition, complicated sample preparation procedures, and long acquisition times, make this system unsuitable (if not incompatible) when attempting live-cell imaging.
In addition, the optical standards and computer power required yield costs prohibitively expensive making SR methods inaccessible to many labs.
Recently, progress with spinning disk confocal technology has proven to be suitable for live-cell imaging achieving a 1.6x improvement beyond the diffraction limit. This was achieved with a combination of optical over-sampling and computational reassignment, however, this came at the cost of a considerably reduced field of view, starting from a decreased native resolution. Besides, it is still inextricably bound to the increased costs of expensive hardware.
The Dragonfly spinning disk confocal has an improved native axial and lateral resolution due to its innovative custom pinhole pitch separation and pinhole size, a combination that delivers improved resolution for routine imaging. Furthermore, utilizing photon reassignment (deconvolution) on the images obtained, the ultimate axial and lateral resolution of the dragonfly can be 240 nm and 139 nm accordingly.
Yet, questions remain such as, what if a researcher requires improved resolution? Or, does not have access to a spinning disk confocal microscope? Is there an alternative method compatible with live imaging, that can rapidly obtain super-resolved images at the full field of view and, if necessary, deep inside the cells? The answer to these questions is absolute when using SRRF or SRRF-Stream+ (Figure 1).
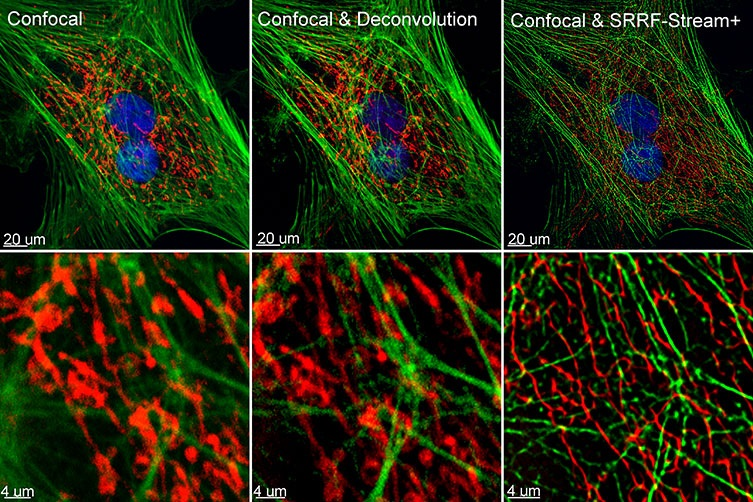
Figure 1. Comparison between confocal imaging, confocal imaging & deconvolution, and confocal imaging & SRRF-stream+. High magnification close-ups are present for better visualization of the obtained resolution. BPA cells stained with phalloidin, moto tracker, and DAP image with Ixon 888 in confocal mode (no SRRF) and confocal mode with SRRF-stream+.
In 2016, the Henriques lab developed an unconventional approach to super-resolution using a technique known as SRRF - Super-Resolution Radial Fluctuations (1). SRRF can be used in combination with widefield, TIRF, or confocal, and the conclusive resolution will rely on the proprieties of the acquired data sets.
Utilizing the SRRF algorithm, the researchers can acquire resolutions in XY up to 50 nm (1). In addition, the SRRF algorithm does not necessitate unconventional or complicated sample preparation or special fluorophores for purchase, making it compatible with standard fluorophores and fluorescent proteins.
Primarily, a super-resolved SRRF image can be obtained by capturing between 20-100 frames on average (more frames result in enhanced resolution), and the energy needed for this imaging is in the order of the mW to W per cm2 range. These properties make SRRF compatible in accordance with live-cell imaging, (1, 2).
Initially, SRRF was offered as an ImageJ plugin, and to acquire an SRRF image, it was necessary for a long acquisition workflow to take place. Therefore, the SRRF-image was eventually obtained by post-acquisition processing of the acquired data in the Image J plugin - NanoJ.
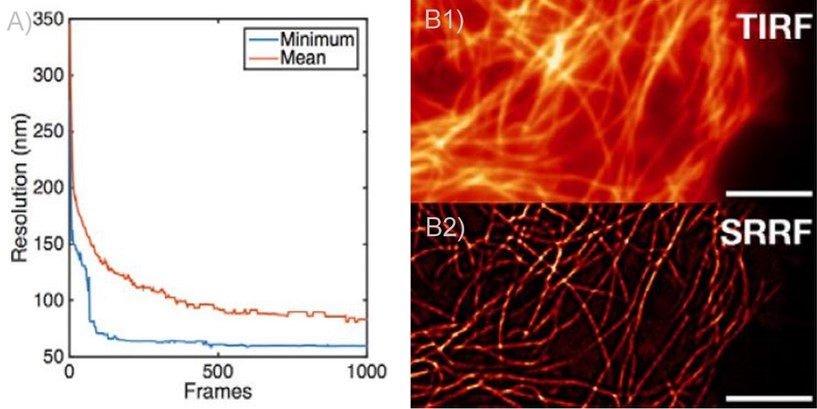
Figure 2. SRRF and SRRF-stream resolution increase with the number of frames acquired (and processed) per time point. A) Graph presenting the resolution increase with the increasing number of acquired frames. As can be observed there is a high increase until 100 frames. Mean presents the average of all radial fluctuation correlation analyzed (in different Regions of interest - ROI). Minimum shows the maximum resolution achieved for a given ROI. B1 and B2) Actin filaments imaged on Andor Dragonfly using TIRF (B1)) and TIRF with on the fly SRRF-stream processing (B2). It can be observed even when acquiring in TIRF modality, that SRRF-Stream provides a significant increase in resolution.
During 2018, a collaboration between Andor and professor Henriques, Andor launched SRRF-Stream which is its application of the SRRF algorithm that generates super-resolution images, in real-time, with the click of a button. As with the initial SRFF algorithm, the resolution of SRRF-Stream will get better with each increasing number of frames taken per a given time point.
This is most prominent up to 100 frames, achieving a more stable increase from 100 frames to 500 frames, and a quite modest improvement in resolution from 500 frames onwards (Figures 2, 3). Notably, other factors will affect resolution such as Nyquist sampling, radiality magnification, exposure time, and the ring radius increase; these should be tested in order to achieve optimal results.
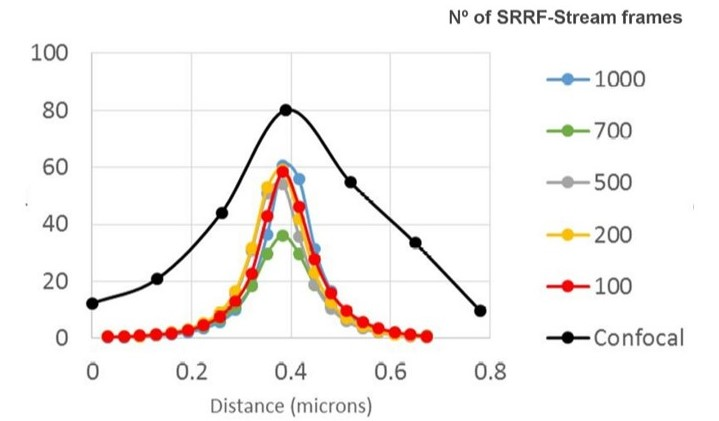
Figure 3. SRRF-stream resolution increase with the number of frames acquired per time point. 100 nm tetraspeck beads where imaged and the FWM was measured. We can observe that with 100 SRRF frames acquired the FMW of the tertaspeck beads is 50 nm. The 100 nm bead can be accurately measured using the SRRF-stream algorithm and acquiring 100 frames. Images were acquired in confocal mode, and in confocal mode with different SRRF-stream acquired image images per point, respectively: 100, 200, 500, 700, 1000. Thanks to Dr. Ann Wheeler, Head of the Advanced Imaging Resource at the MRC Institute of Genetics and Molecular Medicine, University of Edinburgh.
ANDOR´s SRRF-Stream was a significant improvement enabling live-cell super-resolution, deep inside cells and tissues exceeding the top of the coverslip.
SRRF-Stream is offered exclusively with iXon-EMCCD cameras, through micro-manager and Fusion which is the software that controls the Dragonfly confocal. The benefits of SRRF-Stream are as follows (3,4):
- Disrupts the diffraction limit of light by providing Super-Resolved images with 2 to 6-fold increase in the final resolution (50-150 nm), depending on which imaging mode is being utilized (i.e. confocal, widefield, or TIRF) as well as the experimental conditions
- Enables Real-Time super-resolution images by improving the workflow and preventing the need for post-processing.
- Use of Low Excitation power intensities for imaging (mW to W/cm2). This results in higher compatibility with prolonged live-cell observations and minimal impact in cell physiology.
- Compatible with a typical fluorophore and fluorescent proteins
- Simple sample preparation
Therefore, SRRF-stream is an economical solution for any microscope since all of them can be transformed into super-resolution microscopes. SRRF-stream is compatible with any imaging technique, notably confocal, widefield, TIRF and (Figure 4). In July 2020 came the announcement of further improvements to the SRRF-Stream algorithm: SRRF-Stream+, (Figures 1, 4, 5, 6, 7).
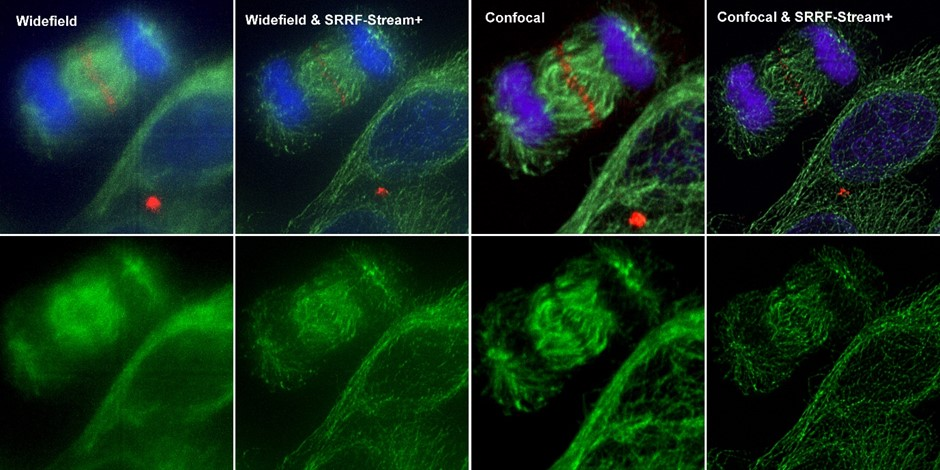
Figure 4. Comparison between different imaging modes (widefield and Confocal) with and without SRRF-Stream+ Hela cells stained for MLKP1(red), a-tubulin(green-microtubules) a and DAPI (blue-DNA) were image in the Dragonfly with an Ixon888 camera.
With enhanced radiality measurements, the SRRF-Stream+ goes a step beyond the previous version of SRRF. Initially, radiality measurements were computed in 6 directions whereas now they are computed in 24 directions (Figure 5). This advancement will refine the results of your super-resolved processing data as well as removing the periodic star artifacts, previously visible specifically in circular structures such as kinetochores (Figure 7).
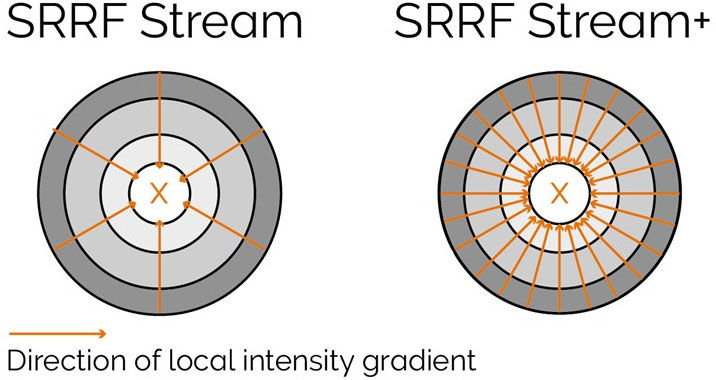
Figure 5. Radiality computation measurements in SRRF-Stream and SRRF-Stream+ As it can be observed by the image the increased computational radiality measurements will deliver a more accurate result of the SRRF image.
To return optimal results from SRRF-Stream+, the user will still need to obtain images under Nyquist criterium. As reported previously, the main advantage in resolution will be acquired when oversampling by a factor of 2.3 or higher. Yet, when oversampling by a factor of 1.5 satisfactory results can still be produced using the SRRF-Stream+.1
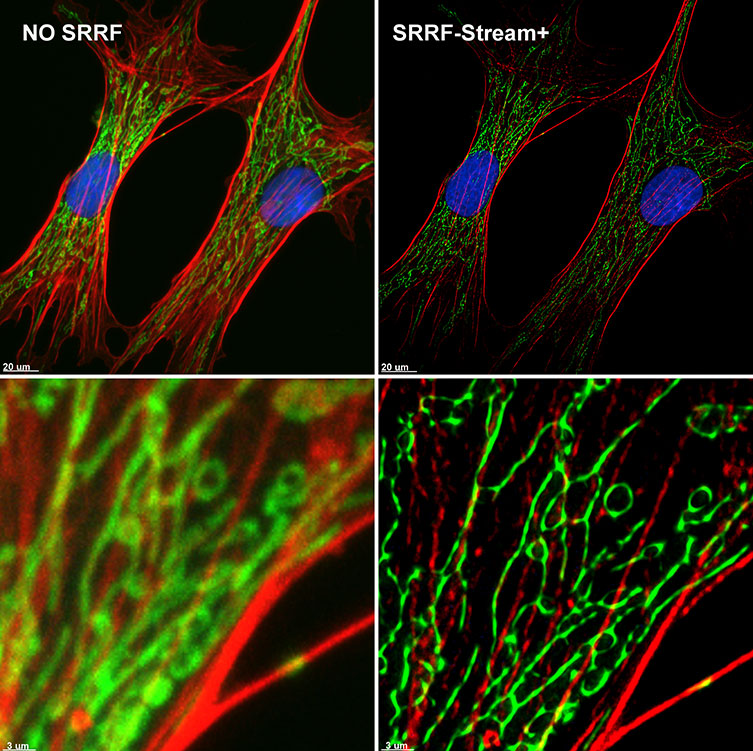
Figure 6. SRRF-Stream+ delivers High-quality Super-resolved images. BPA cells stained with phalloidin (red), mitotracker (green), and DAPI (blue) were imaged with Ixon 888 Ultra in confocal mode (no SRRF) and confocal mode with SRRF-stream+.
When analyzing the illustrations visible in Figures 1, 4, 5, and 7 the advantages of SRRF-Stream+ are clear, but many potential users may still have a few significant questions: how is the speed of acquisition influenced? Is SRRF-Stream+ 4x slower than the original version of SRRF, if SRRF-stream+ computes radialities in 24 direction instead of 6?
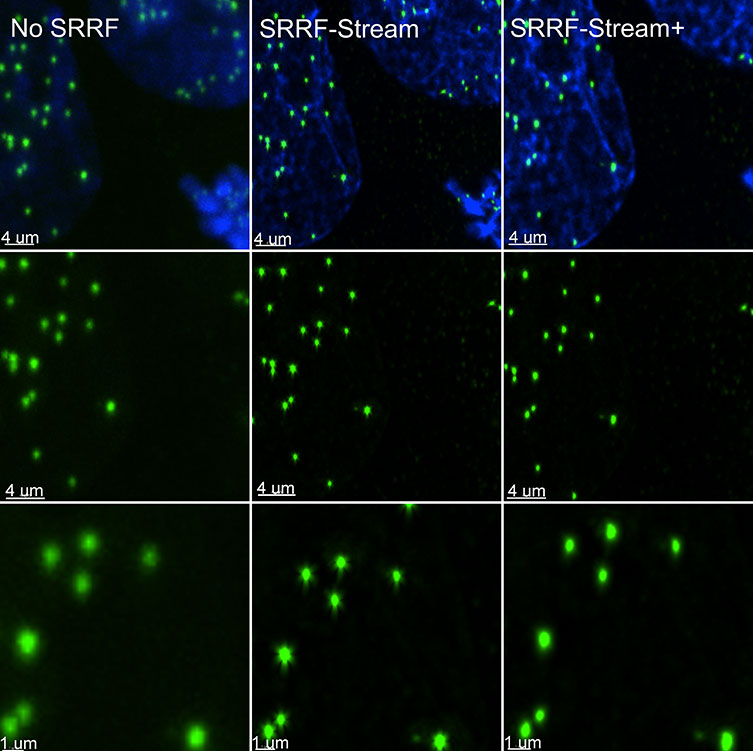
Figure 7. SRRF stream+ delivers high-quality data. At very high magnifications, in specific structures, such has kinetochores some Star artifact could occasionally appear. SRRF-Stream+ radiality computation in 24 directions eliminates the star artifacts. Hela cells stained for Cenp-A (green-Kinetochore) and DAPI (blue-DNA) were imaged in the Dragonfly with an Ixon888 Ultra camera.
When the latest SRRF algorithm was in development, Andor considered that the speed of acquisition is also a crucial parameter to many researchers. It has been possible to obtain enhanced image quality with a nominal impact on acquisition speeds. To accomplish this, CUDA performance has been maximized entirely by utilizing the massive computing power of nVidia graphic card to speed up SRRF-Stream+ calculations.
Consequently, a time-lapse of 100-time points with 50 SRRF-stream+ images per time point (equivalent to 500 images in total) will only differ by 0.5 secs in acquisition comparative to the original SRRF-stream.2
With regards to the benefits of Andor’s original SRRF-Stream, they are preserved in the SRRF-Stream+ but with enhanced image quality. Super-resolution imaging processing is still completed at rapid processing rates, up to 30X faster than ImageJ SRRF (“NanoJ-SRRF’). Furthermore, SRRF image acquisition/processing is captured in real-time, parallel to the procurement of the data: the algorithm generates images with a final resolution between 50-150 nm.
Furthermore, because of the low energy requirements, SRRF-Stream+ is the perfect solution for live-cell microscopy. Super-resolved images can be obtained by utilizing large fields of view, and crucially, super-resolution microscopy is not limited to cell surface events. With SRRF-Stream+, it´s conceivable to gain a super-resolved image deep inside cells or tissues, without the need for complex sample preparation.
Subsequently, since the launch of the SRRF-Stream, it has become extensively used for various applications. Examples of the SRRF-Stream being used in iXon cameras(4) include: to observe mitochondria trafficking into cell terminus via microtubule destabilization(5), or examination of HIF1α nuclear translocation in mesenchymal stem cells.(6)
The Dragonfly multimodal confocal system was also utilized to generate SRRF-Stream images, examples include the study of cytokinesis using optogenetic tools(7) in addition to vesicle trafficking analysis on studying the effects of class II PI3Ks in the regulation of clathrin-dependent pinocytosis.(8)
Additional applications of SRRF-Stream+ involve investigations of protein structures at a sub-organelle level, membrane fusion studies of individual SNARE protein machinery, tracking of single molecules inside cells, and intracellular skeleton reassembly (alterations to actin fiber meshwork).
In conclusion, the advancement of the SRRF-Stream algorithm delivers live-cell compatible super-resolution with significantly improved results. SRRF-Stream+ generates super-resolved images of intracellular structures free of artifacts, compensating for the effects of cameras fixed pattern noise, in turn, providing enhanced, high-quality super-resolved images.
Bibliography
- Gustafsson, N., Culley, S., Ashdown, G. et al. Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations. Nat Commun 7, 12471 (2016). https://doi.org/10.1038/ncomms12471
- Culley S, Tosheva KL, Matos Pereira P, Henriques R. SRRF: Universal live-cell super-resolution microscopy. Int J Biochem Cell Biol. 101:74-79 (2018). https://doi.org/10.1016/j.biocel.2018.05.014
- Browne M. Twelve Reasons Why Your Next Confocal Should Be Dragonfly. (2017) https://andor.oxinst.com/ebooks/why-your-next-confocal-microscope-should-be-dragonfly
- Coates C, ‘SRRF-Stream’: Real-Time Super-Resolution in a Camera. (2017) https://andor.oxinst.com/assets/uploads/documents/srrf-stream-technical-note.pdf
- Choi, G.E., Oh, J.Y., Lee, H.J. et al. Glucocorticoid-mediated ER-mitochondria contacts reduce AMPA receptor and mitochondria trafficking into cell terminus via microtubule destabilization. Cell Death Dis 9, 1137 (2018). https://doi.org/10.1038/s41419-018-1172-y
- Lee, H.J., Jung, Y.H., Oh, J.Y. et al. BICD1 mediates HIF1α nuclear translocation in mesenchymal stem cells during hypoxia adaptation. Cell Death Differ 26, 1716–1734 (2019). https://doi.org/10.1038/s41418-018-0241-1
- Castillo-Badillo J.A., Bandi A. C., Harlalka, and Gautam N. SRRF-Stream Imaging of Optogenetically Controlled Furrow Formation Shows Localized and Coordinated Endocytosis and Exocytosis Mediating Membrane Remodeling. ACS Synth. Biol. 9, 4, 902–919 (2020). https://doi.org/10.1021/acssynbio.9b00521
- Aung, K.T., Yoshioka, K., Aki, S. et al. The class II phosphoinositide 3-kinases PI3K-C2α and PI3K-C2β differentially regulate clathrin-dependent pinocytosis in human vascular endothelial cells. J Physiol Sci 69, 263–280 (2019). https://doi.org/10.1007/s12576-018-0644-2.
References and Further Reading
- For more detailed information on how to sample on Ixon cameras please go to Andor original SRRF-stream tech note.
- Full field of view (1024 X1024) images acquired with Ixon Ultra.

This information has been sourced, reviewed and adapted from materials provided by Andor Technology Ltd.
For more information on this source, please visit Andor Technology Ltd.