Spatial transcriptomics (also known as multiplexing) is a cell biology technique able to uncover a number of (Xn) RNAs in their 2D or 3D biological context. Multiplexing is a popular approach in areas such as oncobiology, neurosciences, disease target diagnostics, development, and behavioral studies.
A number of techniques have been developed to enable spatially resolved transcriptomics - also referred to as multiplexing or spatial genomics. These techniques include FISSEQ, osmFISH, instaSEQ, MERFISH, STARmap and seqFISH.
Spatial transcriptomics offers a number of benefits, including its ability to provide spatial and sequence information for multiple gene products. Until recently, it was only possible to sequence RNAs in bulk, or at the single-cell level, with any tissue environment information (the biological context) lost during this process.
A thorough understanding of where genes are expressed and their surrounding environment (microenvironment) has the potential to uncover the molecular basis of brain function, development, cancer and neurodegenerative diseases.
Multiplexing hybridization experiments generally involve multiple rounds of hybridization and detection. Each of these rounds is executed sequentially in the same sample.
Staining (Xn) molecules require the probes to be removed and the sample hybridized once again. Attaching a microfluidic system to the Dragonfly spinning disk confocal system enables this.
Mechanistically, fluorescent probes will label the hybridized RNA molecules. Image data is acquired – generally by scanning a volumetric montage - before the probes are washed away. After the acquisition of each image dataset, a ‘strip and wash’ step is employed, followed by a further hybridization round.
This procedure will be repeated N times, resulting in substantial volumes of encoded image data. The infographic below summarizes the sequential steps necessary for the acquisition of multiplex imaging using the Dragonfly confocal.
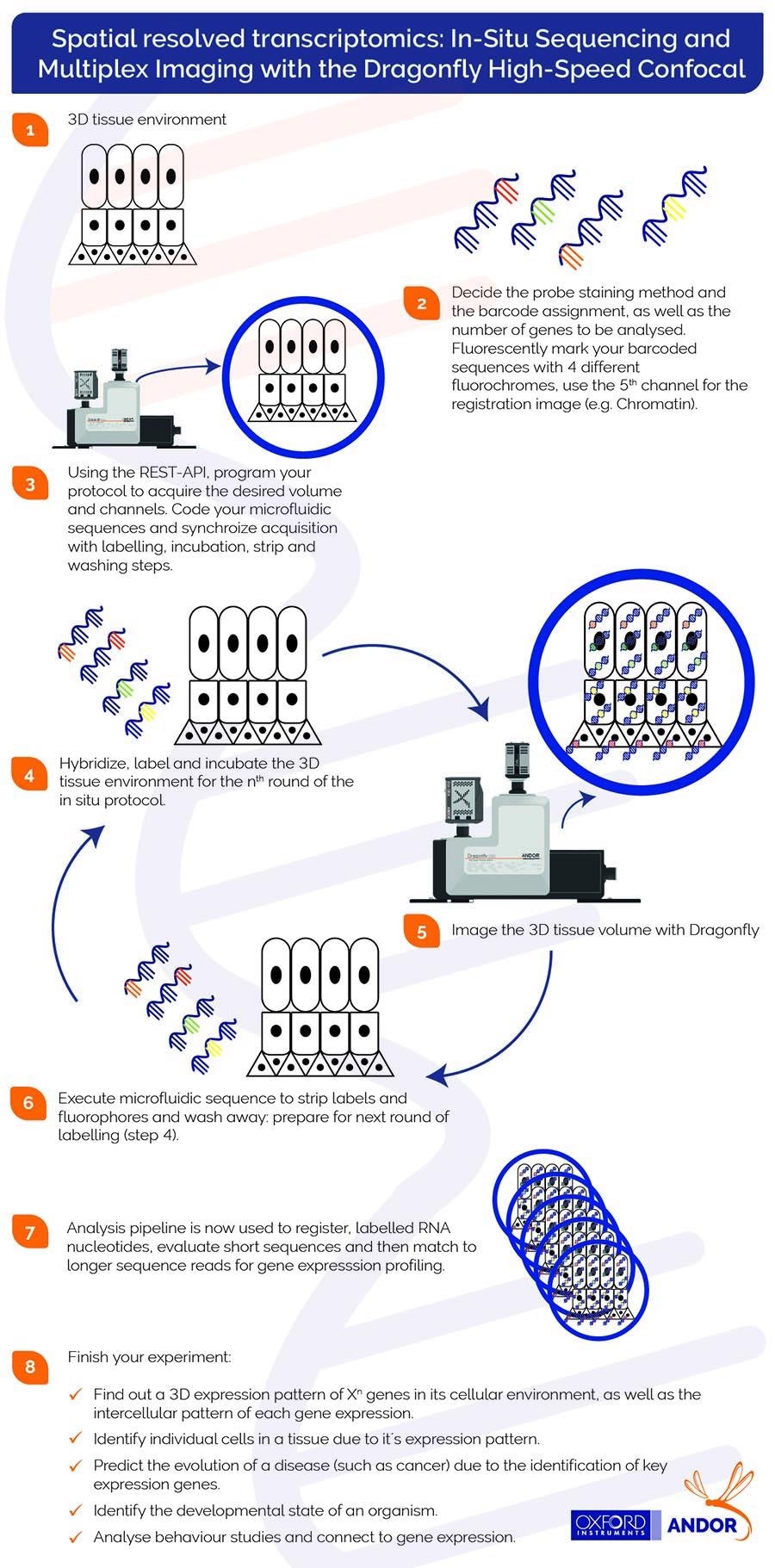
Image Credit: Andor Technology Ltd.

This information has been sourced, reviewed and adapted from materials provided by Andor Technology Ltd.
For more information on this source, please visit Andor Technology Ltd.