In quality control and chemistry, analytical samples are usually mixtures. Understanding the main compound’s assay and the number of impurities or other unwanted substances in the sample is essential for analyzing reaction conditions. However, it is difficult to measure impurities accurately without knowing their identity.
This article demonstrates achieving quantification by assessing the molecular weight with nuclear magnetic resonance (NMR) using the 1H-detected diffusion-ordered spectroscopy (DOSY) experiment.
If 1 kg of pumpkins is required to make pumpkin soup, determining how many pumpkins to purchase is impossible if their weights are unknown. The same issue occurs when handling analytical samples.
In chemical manufacturing, samples frequently comprise unidentified components. NMR spectroscopy is a relative primary quantification technique, and peak integrals can be employed to subtract the molar amount of such components from the total of an identified internal reference standard.
If a peak in the NMR spectrum comes from an unidentified molecule, the gravimetric amount cannot be determined from the molar amount because the molecular mass in g/mol is unspecified.
This article illustrates that estimating the molecular mass from 1H NMR DOSY data is possible, facilitating robust investigational approaches for mixture analysis.
Analyzing Gravimetric Sample Composition for Reaction Conditions
Establishing the gravimetric sample composition is beneficial for characterizing quantitative sample properties, which is crucial in optimizing reaction conditions toward higher yield and purity (see Figure 1).
The gravimetric sample composition determines how much each component of a mixture impacts the total mass; the sum of all elements must be 100 % weight/weight (w/w).
A 10 mg sample of a reaction product, for example, comprises three components:
- 5 mg of product
- 2.5 mg of starting material
- 2.5 mg of side product or residual solvents
The product’s gravimetric share is 50 % w/w, while the starting material and side product form 25 % w/w each.

Figure 1. If the assay of the main compound accounts for 50 % w/w of the sample mass, there is a risk that the remaining 50 % contain potentially harmful substances that remain undetermined (A). Product and impurities make up for 100 % w/w of the sample mass indicating that the sample is well characterized (B). If one compound or the sum of all compounds exceeds 100 % w/w, the results of one or several analytical methods are likely compromised (C). Image Credit: Bruker BioSpin Group
Approaches that can accurately recognize the precise number of elements and consistently calculate the quantity of these elements are needed for this analytical task.
Frequently employed approaches for this task include high-performance liquid chromatography (HPLC) to identify UV active compounds, HPLC-mass spectrometry (MS) to identify ionizable compounds, and gas chromatography to identify volatile compounds.
Other methods specialized in recognizing certain additional substance classes are also frequently utilized. As shown in Figure 2, NMR is significant as it identifies the common 1H nucleus in most organic molecules.
This indicates that NMR is not constrained to similarly small substance classes like UV-active or ionizable compounds. Detecting and calculating chemical compounds is challenging and typically demands numerous analytical methods.
The number of distinct substances existent is frequently unknown. Substances can remain unidentified even if recognized, and analytical approaches can be inaccurate, causing false negatives or positives and incorrect numerical results.

Figure 2. In instrument-based analytics, various solutions are available for different classes of substances, for instance ICP-OES (Inductively Coupled Plasma Optical Emission Spectroscopy), IC (Ion Chromatography), Titration, HPLC, HPLC-MS (HPLC coupled with Mass Spectrometry) or GC. NMR can be used for all substance classes which can be analyzed with the aforementioned methods. Image Credit: Bruker BioSpin Group
The challenges above are typically addressed by developing dedicated and optimized analytical techniques, a time-consuming process often performed throughout product development to determine release testing procedures.
Such enhanced and dedicated analytical techniques do not typically exist in research and development (R&D) or when screening numerous reaction conditions, making chemical development daunting.
Establishing the gravimetric sample composition and knowing that the total of all compounds must equal 100 % w/w makes it easy to verify the validity of the analytical techniques utilized.
Determining the gravimetric sample composition is consequently “good practice” in various fields of analytical chemistry, including in R&D, qualifying reference standards, and the pharmaceutical release process.
When evaluating the quality of an active pharmaceutical ingredient, the gravimetric sample composition is determined according to the active ingredient (the product), starting material, and inorganic residue, as well as impurities, residual solvents, and related compounds, which account for 100 % of the w/w sample mass.
- A sum totaling < 100 % often indicates product contamination via unidentified compounds.
- A sum totaling > 100 % often implies that one or several quantitative results are incorrect.
Gravimetric Sample Composition vs. Molar Sample Composition
The molecular mass in g/mol links the gravimetric sample amount in grams to the amount of substance in mol.
In practice, the gravimetric amount is more beneficial as it can be established with a balance. A substance’s molar amount is typically unknown and can only be established with techniques like NMR.
Analytical Techniques and Their Quantitative Abilities
After establishing the number of substances in a sample and identifying each one, the next step is quantification. In various analytical techniques, quantification involves comparing peak integrals or peak intensities.
Depending on the technique, the challenge is that integrals and intensities frequently rely on the molecule under analysis, making it impossible to assess peaks of one molecular species (e.g., a reference standard) alongside those of another molecular species (e.g., an unknown compound). This is the case for numerous universal techniques, including HPLC and HPLC-MS.
Quantification typically demands knowing the exact molecular identity and availability of a qualified reference standard of the same material. Assuming a sample contains five known molecular species, five separate reference standards are required to quantify.
The availability of reference standards limits characterizing analytical samples and makes quantification long and costly.
NMR spectroscopy, however, is fundamentally quantitative, meaning the integral of an internal reference standard can be assessed alongside a random quantity of distinct molecular species. This makes NMR spectroscopy the optimum approach for consistent and rapid compound quantification in mixtures.
Predefined NMR experiments for identifying common nuclei include 1H, 13C, 23Na, or 35Cl. Many substance classes can accordingly be assessed with NMR:
- 1H: Organic molecules
- 23Na: Various inorganic sodium salts, including NaCl and Na2 SO4
- 35Cl: Various inorganic salts and the substance class of organic hydrochlorides
The gravimetric sample composition of most associated mixtures can therefore be determined by only preparing one NMR sample utilizing the inherent quantitative properties of NMR. The related NMR experiments can be obtained automatically in just several minutes.
The next section of this article demonstrates how to extend this approach to unknown organic substances by approximating the molecular weight in g/mol from 1H NMR data.
Estimating the Molecular Weight of Unspecified Mixture Compounds
DOSY employs spatial encoding in the NMR sample to calculate molecular diffusion directly proportionate to the molecular weight in g/mol, facilitating robust experimental strategies by combining mass data and NMR data.
The DOSY Experiment
The DOSY experiment presents the standard 1D 1H NMR spectrum on the x-axis and matches each peak with a diffusion constant on the y-axis. Signals aligned horizontally are likely from the same molecule, making the DOSY one of the simplest yet most robust NMR techniques.
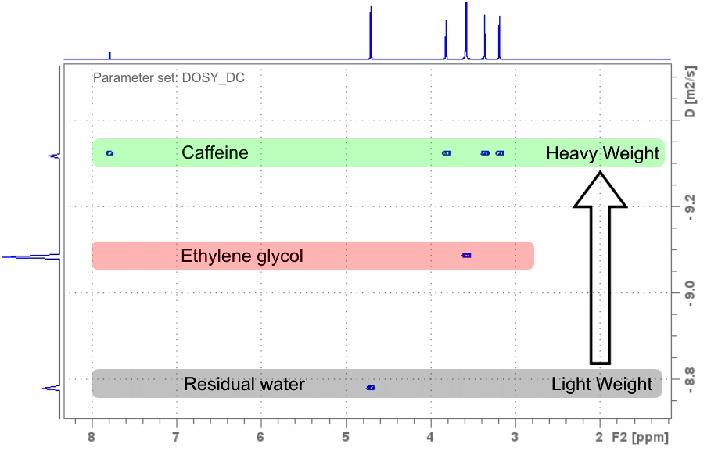
Figure 3. 1H NMR DOSY experiment recorded with the DOSY_DC parameter set in 2:55 minutes on a sample of approx. 5 mg caffeine and ethylene glycol in D2O using an AvanceCore 400 MHz NMR spectrometer. Image Credit: Bruker BioSpin Group
Utilizing calibration data from identified molecular weight standards enables the diffusion constant to be connected with approximate molecular mass in g/mol. This analytical approach is reliable even in adverse conditions, including:
- Unassigned unknown NMR peaks
- Groups of peaks that may or may not derive from the same molecule
The DOSY’s sensitive 1H detection with an extremely high dynamic range and extremely low 2D artifact recognizes and reliably classifies trace compounds when more concentrated substances from the sample matrix appear. This is unparalleled over many other 2D experiments.
A substance can also be recognized in the DOSY spectrum even if just one signal is resolved. This makes the DOSY especially beneficial when trace compounds with bigger molecular weights exist, which could show numerous overlain signals.
Determining Unknown Mixture Composition by Gravimetric Analysis
Sample Preparation
Quinine was selected as the primary compound to prove the hypothesis, and various other organic molecules were combined to mirror impurities.
These impurities included benzene, trimethoxybenzene, MeOH, PEG, and tBuOH. To demonstrate this example, their identity is assumed to be unknown.
Table 1. Test sample composition containing TCNB (1,2,4,5-Tetrachloro-3-nitrobenzene) as a reference standard and quinine to represent the product. Benzene, trimethoxybenzene, MeOH, PEG and tBuOH were added to mimic impurities. The sample was dissolved in deuterated DMSO. The net weights were determined with a balance when the sample was prepared. Source: Bruker BioSpin Group
| |
M
[g/mol] |
Net Weight
[mg] |
Theoretical Gravimetric
Share [% (w/w)] |
| TCNB Reference Standard |
261 |
12.243 |
- |
| Quinine |
324 |
18.042 |
85.4 |
| Benzene |
78 |
0.721 |
3.4 |
| Trimethoxybenzene |
168 |
1.491 |
7.1 |
| MeOH |
32 |
0.447 |
2.1 |
| PEG 10.37 kDA |
10370 |
0.437 |
2.1 |
| tBuOH |
74 |
0.458 |
2.2 |
| Sum |
- |
21.138 |
100.0 |
The theoretical assay (Table 1, column “Theoretical Gravimetric Share”) was established from the volume of a substance by weighing upon test sample preparation.
The theoretical assay for quinine is 85.4 % w/w (18.042 mg / 21.138 mg * 100), meaning around 15 % of the sample mass are impurities that harm product quality.
1H NMR Examination
The 1H nucleus is abundant in most organic molecules. Its high sensitivity makes it the optimum choice for investigating mixtures.

Figure 4. 1H NMR of the quinine sample in deuterated DMSO. Most of the signals originate from the main compound, quinine. Apart from that, signals from a variety of organic impurities (see Table 1) are resolved. Image Credit: Bruker BioSpin Group
Assuming the organic impurities are unidentified, the following step would typically involve 2D experiments like total correlation spectroscopy (TOCSY).
Two-Dimensional 1H NMR Examination
Two-dimensional 1H NMR experiments are highly sensitive and are, therefore, suitable for investigating unidentified mixtures and molecules.

Figure 5. 1H TOCSY spectrum of quinine containing a variety of impurities (Table 1). The traces of the impurity signals are labeled in yellow. They are characterized by the absence of cross peaks. Image Credit: Bruker BioSpin Group
The sensitive TOCSY experiment employs scalar coupling (through-bond coupling) to generate cross peaks and expose the molecular structure. Singlet signals are restricting as they do not demonstrate spin coupling or generate cross peaks.
Singlet signals are common in many small organic molecules and the impurities in the quinine sample. In the TOCSY experiment, these signals, marked in yellow (Figure 5), have no 2D cross peaks. No coupling partners can be established, leaving the number of molecules in the mixture and their structure unidentified.
The following try could involve recording a through-space experiment instead of a through-bond experiment, e.g., Nuclear Overhauser Effect Spectroscopy, to correlate protons with one another through space.
However, the lower signal-to-noise ratio and higher artifact-to-signal ratio render the experiment largely ineffective for assigning unidentified, diluted components in mixtures.
Other heteronuclear NMR experiments face similar issues. As the quantity of molecules in the mixture is unidentified, assigning them with spectral artifacts (false positives) while possibly lacking some signals due to inadequate signal-to-noise (false negatives) would likely prove ineffective.
Recognizing Impurities with the DOSY Experiment
The DOSY and standard 1H experiments can be recorded from the same sample in just a few minutes of experimental time. They can be achieved in manual mode or with automation software like GoScan and IconNMR.
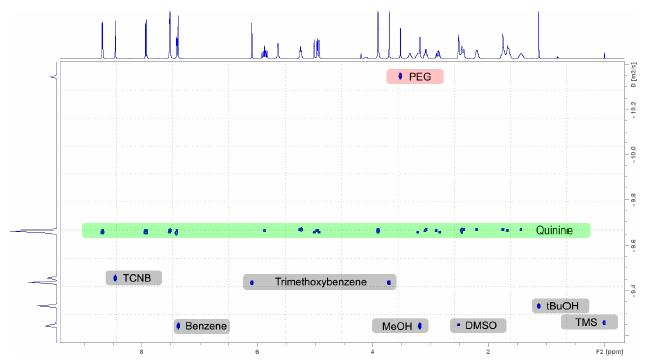
Figure 6. DOSY spectrum of the quinine sample in DMSO containing the reference standard TCNB and the impurities benzene, trimethoxybenzene, MeOH, PEG and tBuOH. Image Credit: Bruker BioSpin Group
Calculating the DOSY scale is as easy as drawing horizontal lines in the 2D plane. The product quinine is the heaviest constituent among the non-polymeric species, and the corresponding signals are labeled in green (Figure 6). This enables fast and consistent assignment even in complicated spectra and even if the quantity of constituents is undetermined.
The impurities, resolved at lower molecular weights (Figure 6, grey rectangles), imply that these signals do not belong and are not derivatives or isomers of quinine.
Similarly, polyethylene glycol’s polymeric nature gives it the largest molecular weight, and its signal is located in the upper section of the 2D spectrum (Figure 6, red).

Figure 7. To unfold the 1H spectrum of the mixture, 1D projections are taken from the DOSY spectrum. The dispersion in the diffusion dimension allows for the resolution of individual components and reaches its resolution limit for benzene, MeOH, DMSO and TMS due to their similar molecular weights. Image Credit: Bruker BioSpin Group
To further examine the mixture, horizontal slices can be obtained from the DOSY spectrum (Figure 7), each of which is a 1D 1H spectrum of the equivalent substance.
The DOSY experiment can acquire data that would have otherwise needed chromatographic sample separation and then the attainment of individual 1H NMR spectra.
Determining the Gravimetric Sample Composition
Quantifying the primary compound is the foremost and most valuable step in determining the gravimetric sample composition. This was achieved by adding the reference standard TCNB (see Table 1) and acquiring a quantitative NMR spectrum (Figure 4) for the test sample.
The spectra displayed in this example were obtained with a 400 MHz AvanceCore NMR spectrometer.

Figure 8. qNMR workflow with the command “nmrq” to calculate the percentage of quinine in a sample of 21.138 mg using 12.243 mg of the reference standard TCNB. The assay of quinine is calculated to be 84.999 % w/w. Image Credit: Bruker BioSpin Group
Assessing the qNMR spectrum:
- Combine the signal of the reference standard (TCNB) with one of the signals of the product (quinine).
- Enter “nmrq” in the TopSpin command line to open the “NMR Quantitative Analysis” window.
- Import the integrals by clicking the import button (Figure 8, No. 3).
- Click the qNMR setup button (No. 4). Complete the matrix with the data for quinine and TCNB.
- Click the calculate button (No. 5). The qNMR result table is displayed (No. 6).
The analytical data now known about the sample is displayed in Table 2:
Table 2. The gravimetric share of the product quinine was found with qNMR to be 85 % of the sample mass. Hence, the remaining 15 % of the sample mass is unexplained by the assay value. Source: Bruker BioSpin Group
| |
M
[g/mol] |
Net Weight
[mg] |
Theoretical Gravimetric
Share [% (w/w)] |
Gravimetric Share
by qNMR [% (w/w)] |
| TCNB Reference Standard |
261 |
12.243 |
- |
- |
| Quinine |
324 |
18.042 |
85.4 |
85.0 |
In R&D, most compounds start as crude mixtures holding numerous compounds like isomers, reagents, solvents, and other byproducts while the product typically represents less than 100 % w/w of the sample mass. Understanding the impurity profile is therefore vital to improving reaction conditions.
This is ideally achieved from the same crude sample minus chromatographic separation and purification of the unknowns.
The DOSY experiment can show its strengths here. As previously mentioned, the DOSY experiment (Figure 6, y-axis)’s diffusion axis displays characteristics like a chromatographic separation, meaning the y-axis presents a dispersion like in an HPLC chromatogram.
This dispersion is employed to “unfold” the 1H spectrum into its molecular components (Figure 7), which derives from the individual interactions between the constituents and the solvent, restricting diffusion to a specific value relative to the molecular weight in g/mol. The DOSY experiment unfolds the 1H spectrum and achieves this like a mass spectrometer.
The DOSY experiment is utilized in the following example to highlight the outstanding 15 % of the sample: Reading the diffusion constant from the y-axis of the DOSY spectrum (Figure 6) and employing known elements like quinine and TCNB for calibration, the molecular weight of the unspecified impurities can be determined via a spreadsheet.
Table 3. Approximate molecular weights in g/mol are given in the column M DOSY. The molecular weights were calculated from the diffusion constants that are found on the y-axis of Figure 6. The deviation from the known molecular weight M in g/mol is given in the column %Deviation. Source: Bruker BioSpin Group
| |
M
[g/mol] |
M DOSY |
| [g/mol] |
%Deviation |
| TCNB Reference Standard |
261 |
- |
- |
| Quinine |
324 |
323 |
0 |
| Unidentified Substance 1 |
78 |
46 |
-41 |
| Unidentified Substance 2 |
168 |
105 |
-37 |
| Unidentified Substance 3 |
32 |
43 |
34 |
| Unidentified Substance 4 |
10370 |
8812 |
-15 |
| Unidentified Substance 5 |
74 |
63 |
-15 |

Figure 9. Refinement of the qNMR evaluation (Figure 8) with respect to the impurities. The calculated molecular weights are entered (1) from Table 3 (column “M DOSY”) and the percentage of each impurity with respect to the sample weight (21.138 mg) is calculated as individual assay values. The molecular weight of the repetitive PEG unit is determined from the DOSY experiment to be approximately 8812 g/mol / 236 units = 37.4 g/mol per repetitive unit. Image Credit: Bruker BioSpin Group
Based on the calculated molecular weights, the assay values for the unidentified impurities can be estimated as Assay in % w/w (Figure 9), achieved automatically by the “nmrq” command. The assay values are added to Table 4 (column “Gravimetric Share DOSY, qNMR”).
Table 4. Amounts of substances are calculated from qNMR using the molecular weight (column M DOSY) obtained from the DOSY experiment. Summing over the signals of the unidentified substances 1 to 5 increases the known gravimetric share from 85 % w/w to 95.3 % w/w. Source: Bruker BioSpin Group
| txt |
M DOSY
[g/mol] |
Gravimetric Share,
DOSY, qNMR [% (w/w)] |
| TCNB Reference Standard |
- |
- |
| Quinine |
323 |
85.0 |
| Unidentified Substance 1 |
46 |
1.8 |
| Unidentified Substance 2 |
105 |
4.4 |
| Unidentified Substance 3 |
43 |
2.9 |
| Unidentified Substance 4 |
8812 |
1.1 |
| Unidentified Substance 5 |
63 |
1.8 |
| Sum |
- |
95.3 |
Summing over the “Gravimetric Share DOSY” column demonstrates that the qNMR analysis now represents an overall mass contingent of 95.3 % w/w, approximately 10 % higher when only considering quinine’s assay value (85.0 % w/w).
At this stage, the unidentified constituents can often be allocated. This may be likely as the chromatographic dimension of the DOSY (y-axis, Figure 6) uncovers the number of constituents. The assignment becomes feasible by understanding the chemical shift, reaction conditions, and chemicals used in the reaction.
Table 5. With the knowledge of the reaction conditions and the NMR data, unidentified substances might be assigned. For example, the unidentified substances 1 to 5 are now assigned to Benzene, Trimethoxybenzene, MeOH, PEG and tBuOH. Using the true theoretical molecular weight (column M), the qNMR evaluation can be repeated, assigning 98.6 % w/w of the gravimetrical share. Source: Bruker BioSpin Group
| |
Gravimetric Share |
| |
M
[g/mol] |
Net Weight
[mg] |
Theoretical
[% (w/w)] |
Assigned, qNMR
[% (w/w)] |
| TCNB Reference Standard |
261 |
12.243 |
- |
- |
| Quinine |
324 |
18.042 |
85.4 |
85.0 |
| Benzene |
78 |
0.721 |
3.4 |
3.1 |
| Trimethoxybenzene |
168 |
1.491 |
7.1 |
7.0 |
| MeOH |
32 |
0.447 |
2.1 |
2.2 |
| PEG 10.37 kDA |
10370 |
0.437 |
2.1 |
1.3 |
| tBuOH |
74 |
0.458 |
2.2 |
2.1 |
| Sum |
- |
21.138 |
100.0 |
98.6 |

Figure 10. In the course of the analysis different shares of the overall gravimetric sample composition are elucidated. Using qNMR, the assay of the main compound is determined to be 85.0 % w/w (Table 2) leaving 15.0 % w/w of the sample mass undiscovered (A). Using DOSY data, the amount of unassigned impurities is determined. This increases the known mass contingent to 95.3 % w/w (B). After assigning the unknown impurities, the known mass contingent increases to 98.6 % w/w (C). Finally, 98.6 % w/w of the overall gravimetric sample composition are assigned which is sufficiently close to 100.0 % w/w to deduce that the sample is well characterized. Image Credit: Bruker BioSpin Group
By fully understanding the identity of the impurities, the precise molecular masses can be determined from the sum formulas to repeat the qNMR calculation as presented in Figure 9.
The subsequent assay values are displayed in the “Gravimetric Share Assigned, qNMR” column (Table 5). Summing over all constituents shows that the qNMR experiment now represents 98.6 % w/w of the sample mass, leaving just 1.4 % unsolved.
This 1.4 % distribution among six qNMR determinations triggers an error of approximately 0.2 % per assay determination, well within typical errors of quantitative NMR.
Therefore, the gravimetric sample composition explains nearly 100 % w/w of the sample mass and establishes the legitimacy of the different assay determinations (Figure 10).
qNMR measurements can also be employed to cross-check the credibility of other analytical results, including gas chromatography, water determination by Karl Fischer, etc., by summarizing each constituent and determining the gravimetric sample composition.

This information has been sourced, reviewed and adapted from materials provided by Bruker BioSpin Group.
For more information on this source, please visit Bruker BioSpin Group.