A heterodimeric protein complex (HPC) derived from Pyrococcus furiosus is recognized for its functional role in RNA binding. According to electron microscopy observation, the total molecular weight is 65 kDa for HPC, which is susceptible to create fibril materials in vitro.
An analysis was performed to characterize homogeneity and stability of HPC utilizing the Wyatt DynaPro® dynamic light scattering (DLS) system under many different buffer conditions. Observations revealed that the pH of the buffer solutions had an influence on the homogeneity and stability of the sample.
Instrumentation
The Wyatt DynaPro DLS system coupled with Dynamics® V7 was used to acquire the DLS data at 20 °C, and Dynamics® V7.0.0.95 was used to perform the analysis. The size distribution of the sample can be directly measured from the intensity auto correlation function, as shown in Figure 1. The polydispersity index provides a direct indicator of the heterogeneity of the each molecular species in the sample.
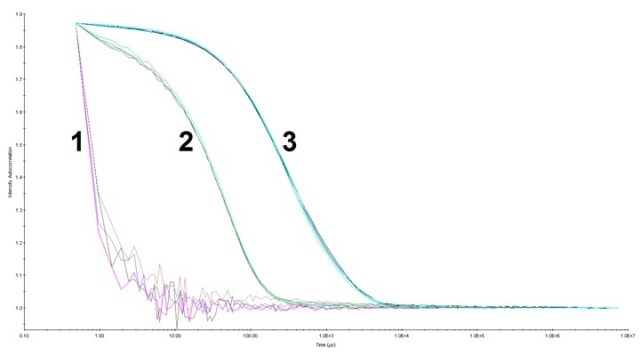
Figure 1. The auto correlation function for HPC at pH 3.0 and pH 7.5. Data was collected with a 5 seconds acquisition time, 10 acquisitions per measurement at laser power 100% (buffer) and 90% (HPC samples). 1) Buffer at pH 3.0 and pH 7.5 (overlapped), 2) HPC at pH 7.5, and 3) HPC at pH 3.0. There was more HPC with a longer decay rate at pH 3.0 (up to ~1.0 x 104 µs) than that at pH 7.5 (up to ~2.0 x 102 µs).
Analysis Results
If a polydispersity index is less than 20%, then the molecular species is mono-disperse, or else it is heterogeneous. The results of the analysis demonstrated that the weighted average polydispersity (%Pd) of HPC at pH 7.5 (Tris-Cl buffer, 3% NaCl) is 19.5%, meaning that it is less polydisperse when compared with HPC at pH 3.0 (Glycine-HCl buffer, 3% NaCl), which has a value of 27.2%.
A total of 10 measurements were carried out for each sample. HPC at pH 3.0 is composed of larger species when compared to that at pH 7.5, which was specified by the longer decay rate in the auto correlation function. As shown in Figure 2, the histogram from the regularization fit of the auto correlation depicts the size distribution of HPC at the aforementioned conditions.
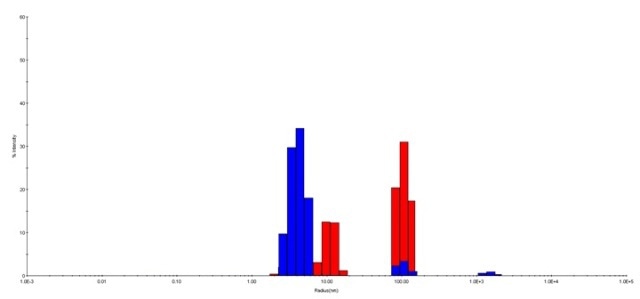
Figure 2. The regularization histograms for HPC at pH 3.0 and pH 7.5. HPC at pH 7.5 (blue) and at pH 3.0 (red) are shown in this histogram. The distribution suggested possible aggregation of HPC occurred at pH 3.0. HPC at pH 7.5 is less polydisperse than that at pH 3.0, with fewer intensive peaks.
From the auto correlation function, it was observed that HPC at pH 7.5 yielded one strong peak around a lower hydrodynamic radius of 4.0 nm, with 90.5% of average peak percentage intensity (average %I), as shown in the size distribution histogram. HPC at pH 3.0 contributed two peaks at a higher hydrodynamic radius of 13.0 nm and 112.4 nm, where the average %I was 32.8% and 64.0%, respectively.
Results suggest that the stability and homogeneity of HPC at pH 7.5 are better when compared to HPC at pH 3.0, because the peak for HPC at pH 7.5 is likely to split into two peaks, which then shift toward higher hydrodynamic radii.
Conclusion
The homogeneity and stability measured from polydispersity and auto correlation function utilizing DLS yield data on the biophysical property of HPC. The study results help corroborate the conditions under which HPC polymerization is thermodynamically more favorable, thereby enhancing the sample preparation quality utilizing suitable buffer conditions for biophysical studies, such as structural analysis.

This information has been sourced, reviewed and adapted from materials provided by Wyatt Technology.
For more information on this source, please visit Wyatt Technology.