Therapeutic recombinant antibodies, principally of the Immunoglobulin G isotype (IgG), account for a growing share of biopharmaceuticals, and a growing number of approved antibody therapies are available in the market for treatment of diseases such as diabetes, rheumatoid arthritis and cancer.
Moreover, many more candidate IgGs are in the biopharmaceutical development pipeline. Hence, gaining more knowledge about these candidates becomes highly significant for successful development. The molecular structure of an example Immunoglobulin is depicted in Figure 1.
Figure 1. Molecular structure of an Immunoglobulin.
Instrumentation
However, the biopharmaceutical industry faces a key problem in the form of protein aggregation. Proteins have the tendency to aggregate over time as they only have a limited shelf-life. Protein aggregates present in the bloodstream can regularly promote the active/adaptive immune response. Size-exclusion chromatography (SEC) is a robust technique for the evaluation of the aggregation content of protein formulations, separating proteins by size to measure their molecular weight and determine aggregate proportions in samples.
When SEC is coupled with a light scattering detector, molecular weights of proteins can be directly measured. Moreover, the setup enables differentiation of monomer, dimer and aggregate irrespective of retention volume. If the light scattering detector used is a multi angle light scattering (MALS), then radius of gyration (Rg) can be measured for the large aggregates which scatter light anisotropically, thus delivering more information about structure and shape.

Figure 2. The Viscotek SEC-MALS 20 instrument.
One such instrument shown in Figure 2 is the Viscotek SEC-MALS 20 system, a 20 angle light scattering device that can make molecular weight measurements of protein and protein aggregates independent of elution volume. This article discusses the separation of a purified polyclonal antibody (IgG) by means of SEC, followed by subsequent characterization utilizing the Viscotek SEC-MALS 20.
Experimental Procedure
Using the TDA RI detector, the SEC-MALS 20 detector was linked to a Viscotek TDAmax system for concentration measurement. Samples were isolated along 2 x Viscotek protein columns. Phosphate buffered saline was used as the mobile phase, where the IgG sample was prepared. The detectors and columns were maintained at 30 °C to achieve good separation and to optimize baseline stability.
Experimental Results
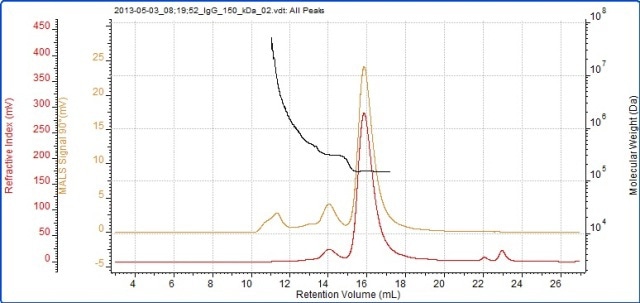
Figure 3. Chromatogram of IgG showing the refractive index (red) and MALS (90°) (orange) detector signals. The molecular weight measured by MALS is overlaid in black.
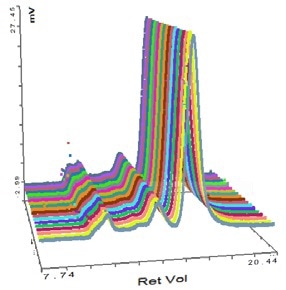
Figure 4. Overlay of MALS detector responses for IgG.
The successful characterization of the IgG sample using the Viscotek SEC-MALS 20 is demonstrated in Figure 3. The data from the different MALS angles is illustrated in Figure 3. The data from the different MALS angles is demonstrated in Figure 4. The calculated molecular weights are listed in Table 1:
Table 1. Measured molecular weights of the different peaks in the IgG sample.
| |
Aggregates |
Dimer |
Monomer |
| Peak RV - (ml) |
13.23 |
14.00 |
15.80 |
| Mn - (kDa) |
674.12 |
308.6 |
147.2 |
| Mw - (kDa) |
7661.00 |
309.2 |
147.4 |
| Mw / Mn |
11.364 |
1.002 |
1.001 |
| Rg(w) - (nm) |
26.6 |
N/C |
N/C |
| Wt Fr (Peak) |
0.014 |
0.065 |
0.921 |
The results reveal that the sample analyzed is actually quite complex and has been isolated into three peaks. The primary peak shows a measured molecular weight of 147 kDa, which is very near to the molecular weight of IgG, indicating that this sample is an IgG monomer.
The secondary peak shows a measured molecular weight of just above 300 kDa, suggesting that the sample is a dimer. Moreover, the molecular weight is very stable across both of these peaks, revealing that they are monodisperse and have well-defined molecular weight and structure.
The third peak is somewhat different, with a light scattering signal far greater than the refractive index signal and with a correspondingly high molecular weight. There is a high variation in the molecular weight, ranging from roughly 600 kDa up to 7x104 kDa. As illustrated by the polydispersity value (Mw/Mn), the third peak is very polydisperse, meaning that it has a very broad range of molecular weights which suggests that it is a non-specific aggregation. Hence, any IgG molecules in these aggregates will probably have lost the majority or all of their activity.
The concentration detector enables quantification of the individual populations. The proportions of these individual populations are also listed in Table 1. The monomer peak consists of 92% of the sample, while the dimer accounts for just 7% of the sample and the large aggregates represent below 2% of the sample.
Proteins are comparatively tiny molecules by the standards of SEC and MALS. Since soluble proteins are isotropic scatterers and normally have a radius of below 10 nm, it is not possible to measure their Rg using MALS. This is correct for the IgG monomeric and dimeric forms analyzed here. However, the Rg value of some of the largest aggregates can be measured, as they have the adequate size to be anisotropic scatterers.
Some angular dependence can be seen in the scattering of the aggregate peak when analyzing Figure 4, providing an average Rg of 26.6 nm. A corresponding Zimm plot obtained from the aggregate peak top is depicted in Figure 5. A Zimm plot taken from the top of the monomer peak, illustrated in Figure 6, shows that the IgG monomer is an isotropic scatterer.
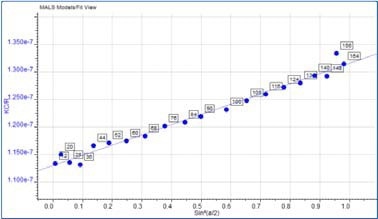
Figure 5. Zimm plot showing the anisotropic scattering for the aggregate peak.
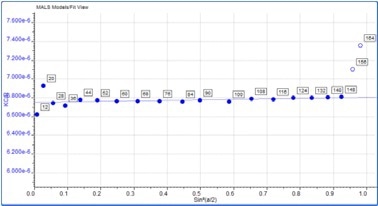
Figure 6. Zimm plot showing isotropic scattering for the monomer peak.
Conclusion
The results clearly demonstrated the ability of the Viscotek SEC-MALS system to measure the molecular weight of proteins independent of their structure and elution volume as well as the molecular size in the form of Rg for molecules that are large enough to demonstrate anisotropic scattering.
The SEC-MALS 20 is an ideal additional detector for any SEC system to provide a tool with a high quality MALS capability that can directly measure protein molecular weights.

This information has been sourced, reviewed and adapted from materials provided by Malvern Panalytical.
For more information on this source, please visit Malvern Panalytical.