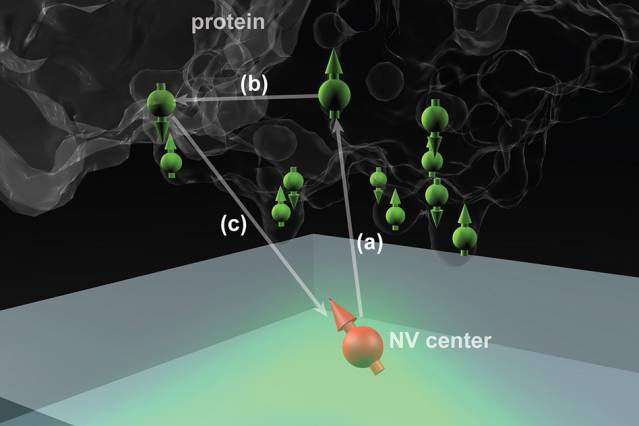
 Nitrogen vacancy (NV) centers in diamond could potentially determine the structure of single protein molecules at room temperature. Here the NV center is 2 to 3 nanometers below the surface, and the protein molecule is placed above it. (Courtesy of the researchers).
Nitrogen vacancy (NV) centers in diamond could potentially determine the structure of single protein molecules at room temperature. Here the NV center is 2 to 3 nanometers below the surface, and the protein molecule is placed above it. (Courtesy of the researchers).
MIT researchers have developed a new technique capable of producing highly detailed images of single proteins irrespective of the level of complexity in the structure and, more importantly, without the need for crystallisation.
Proteins form the building blocks of all living things on Earth. There are almost an unlimited number of protein varieties in existence but as most are highly complex structures, only a small amount of proteins have so far been determined. Determining the structures of proteins is very important when studying basic biological processes and for the development of new drugs.
Conventionally, in order to examine the atomic arrangement of a protein in detail it needs to be first crystallised. This crystallisation process is a difficult task to perform and in some cases it is impossible.
A team of researchers at MIT, comprising of MIT graduate student Ashok Ajoy, postdoc Ulf Bissbort and associate professor of nuclear science and engineering Paola Cappellaro, in conjunction with researchers from the Harvard University and the Singapore University of Technology and Design, have proposed a new method to address this issue.
Microscopic defects which can be introduced in a controlled manner within a diamond’s crystal structure form the basis of the new technique developed at MIT. These nitrogen-vacancy (NV) centres form during the introduction of nitrogen atoms into the crystal structure to replace a carbon atom in a diamond lattice with perfect spacing. These lattices may also have naturally occurring vacancies, which are defects caused by the absence of a carbon atom from its original position within the lattice.
The combination of a nitrogen atom and a vacancy results in the formation of NV centres, which can be used in order to determine the attributes and position of electrons and protons in the atoms around them.
The NV centres emit fluorescence when the diamond surface is irradiated by laser light. The subsequent detection and analysis of the emitted light can provide details of the spin states of nearby particles.
Several research teams are attempting to harness these NV centres for quantum communication and quantum computing applications. If the distance between the NV centres and the surface of the diamond is only a few nanometres, these centres can be used to detect the spin states of particles inside a molecule located on the diamond surface. In turn, this allows the molecular structure to be determined by detecting and mapping out single atoms and their relative positions.
Ajoy explained, “The idea is to place a biological molecule on top of the diamond, and try to determine its structure… With proteins, the structure and function are closely related,” Precisely mapping out the structure helps to understanding basic biological processes and develop new drugs for interaction with specific molecular targets.
Ajoy went on to say, “It could help in developing something that fits on or around [a target molecule], or blocks it, and the first step is to know the structure.”
Nuclear magnetic resonance, transmission electron microscopy and X-ray crystallography are widely used techniques to reveal the molecular structure of proteins. The need for a large sample volume is an issue for all of these techniques, thus limiting their applicability in the analysis of single molecules.
“There are many molecules where this doesn’t work out, because you can’t grow the crystals, or they are very hard to grow,” Ajoy said. “For these molecules, our method might be useful because you don’t need the crystals, you just need a single molecule… the new technique perhaps can determine structure at room temperature, under ambient conditions,”
This research, published in the Physical Review X journal, is currently in the theoretical stage and the researchers at MIT are now working on generating actual images using this technique. “We started building this setup a year ago, and we have preliminary experiments,” Ajoy says.