Mar 28 2016
Infections with bacterium Clostridium difficile have rapidly become a significant medical problem in hospitals and long-term care facilities. The bacteria cause diarrhea and life-threatening inflammation of the colon by producing toxins that kill the endothelial cells that form the lining of the gut.
Although a natural inhibitor of these toxins, called InsP6, works in the test tube, it is not very efficient when administered orally. Traditional methods to optimize InsP6 have until now not been successful, but researchers at Baylor College of Medicine have discovered that changing one atom in InsP6 can increase its ability to neutralize the toxins by 26-fold. The results appear in Science Advances.
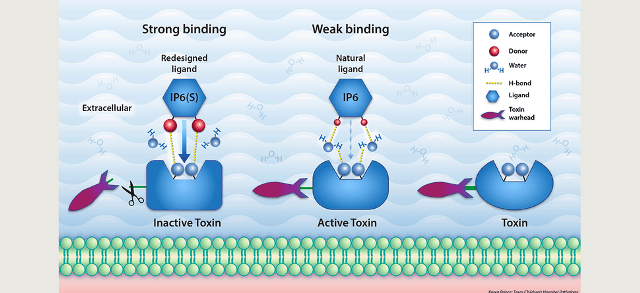 IP6, the natural inhibitor or ligand of Clostridium difficile’s toxin, binds weakly to the toxin. This results in the toxin remaining active (center figure). Changing a single atom in IP6 to form IP6(S) results in stronger binding to the toxin and its inactivation (left side figure).
IP6, the natural inhibitor or ligand of Clostridium difficile’s toxin, binds weakly to the toxin. This results in the toxin remaining active (center figure). Changing a single atom in IP6 to form IP6(S) results in stronger binding to the toxin and its inactivation (left side figure).
“The toxins, called TcdA and TcdB, are very large molecules that kill the cells very efficiently,” said Dr. Tor Savidge, associate professor of pathology and immunology and of pediatrics, director of the Savidge Lab at the Texas Children's Microbiome Center and senior author of the paper. “It’s like delivering a warhead into the cell. The toxins bind to the cell and the cell internalizes them in a sack of cell membrane called endosome. Not all of the toxin will exit this sack to kill the cell, just the little warhead pokes its head out. Another section of the toxin senses when the warhead is outside the sack and cleaves it. The warhead is released, interferes with basic functions and kills the cell,” said Savidge.
To neutralize the toxins, the researchers targeted the section that senses when the warhead is inside the cell, called allosteric modulator. “The strategy we have tried is to make the toxin ‘think,’ before it binds to and enters the cell, that the warhead is ready to be released, so it releases it prematurely,” said Savidge. When the warhead is released outside the cell, it is neutralized. InsP6, the toxins’ natural inhibitor, works this way, but is not very efficient.
Finding molecules that would bind to the allosteric modulator and trigger the premature release of the warhead involved analyzing and testing half a million molecules listed in large databases. Dr. Numan Oezguen, a member of the Savidge Lab, used virtual drug screening to sift through the databases to identify candidate molecules that most likely would bind to the allosteric modulator. One of his screening strategies consists of creating virtual 3-D structures of the molecules, projecting them on a large screen and using 3-D glasses to determine the most likely interactions between molecules. The molecules whose virtual analysis suggested they would bind to the allosteric modulator were then tested in the lab.
“We found that allosteric mechanisms are very complicated,” said Savidge. “You can find something that binds and you think, well, this is probably a good candidate for this, but it’s not right. It binds, but it doesn’t trigger the premature release of the warhead.”
Far from discouraging their efforts, the results motivated the researchers to better understand what makes interactions between molecules stronger or weaker. Their comprehensive analysis of numerous molecules provided insights into how water contributes to molecular interactions, in particular those involving hydrogen bonds, one of the most important bonds between molecules. The roles of water and hydrogen bonding had not been considered in this way before.
“When you take water into consideration you need to acknowledge that it can form hydrogen bonds, which may or may not compete or interfere with those formed between other molecules such as C. difficile toxins and their inhibitors, which interact in the gut, surrounded by water,” said Savidge.
“Before we considered the role of water, the predominant idea was that to strengthen the interaction between molecules the ability to form hydrogen bonds had to be made as strong as possible in the drug. It turns out this is not the case,” said Oezguen. Many times drugs designed to be able to make strong hydrogen bonds bind poorly to their targets.
The researchers discovered that to enhance the binding of a drug to its target, both sides of the hydrogen bond, the side on the drug and the one on the target, have to have either significantly stronger or significantly weaker hydrogen bonding capabilities. On the other hand, a mixed strong-weak hydrogen bond pairing decreases the overall binding of the drug to its target, in some cases by 3 million fold. The decrease in binding is the result, the researchers propose, of water molecules forming hydrogen bonds with the drug and its target, therefore preventing the drug and the target from forming hydrogen bonds between them.
With all this information in hand, the researchers proceeded to make chemical changes in the InsP6 inhibitor to improve its hydrogen bonding capabilities with C. difficile toxins. One of the modifications, changing one single atom in InsP6, strengthened InsP6 binding to the allosteric modulator by 26-fold. This observation builds on a report published by Savidge in Science last year exploring the role of water interactions in the origin of enzymatic catalytic power. Plans are currently underway to exploit these fundamentally new concepts in the precision design of future therapeutic applications.
Other contributors to this work are Deliang Chen, now in Gannan Normal University, China, who contributed conceptual and theoretical proof of the hydrogen bonding pairing principle; Petri Urvil from Baylor performed lab studies; Colin Ferguson from Echelon Biosciences, Inc. synthesized the more efficient inhibitor; and Sara Dann from the University of Texas Medical Branch in Galveston, contributed animal studies.
This work was supported by grants RO1AI100914 and DK56338 from the National Institute of Allergy and Infectious Diseases and the National Institute of Diabetes and Digestive and Kidney Diseases at the NIH, and the National Science Foundation of China (21473041).