Aug 21 2019
Site-directed spin labeling (SDSL) used along with electron paramagnetic resonance (EPR) spectroscopy has been a reliable method for throwing light on the dynamics, structure, and function of proteins and protein complexes.
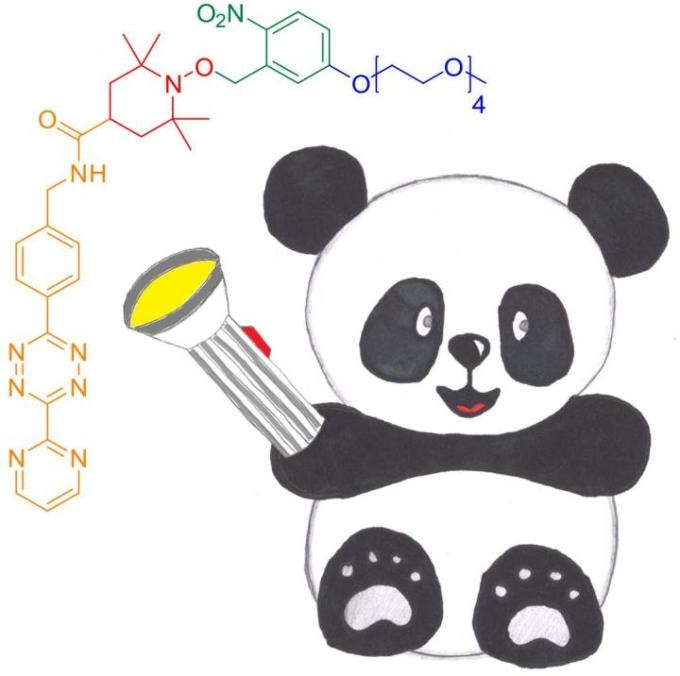 The novel photoactivatable nitroxide for DAinv reaction spin label for proteins, PaNDA. It can be ligated to proteins through a DAinv cycloaddition to genetically encoded noncanonical amino acids. (Image credit: Anandi Kugele)
The novel photoactivatable nitroxide for DAinv reaction spin label for proteins, PaNDA. It can be ligated to proteins through a DAinv cycloaddition to genetically encoded noncanonical amino acids. (Image credit: Anandi Kugele)
Nitroxide-based spin labels are among the most widely used and well-established ones as they are small, non-disturbing, and display outstanding spectroscopic properties.
“Ideal spin labelling procedures exhibit high reaction rates and selectivity,” explained Professor Malte Drescher, Professor for Spectroscopy of Complex Systems at the University of Konstanz’s Department of Chemistry and the study’s main author together with Professor Valentin Wittmann, who is an expert in organic synthesis.
Achieving high reactivity and high selectivity both at the same time can be a problem. Conventional spin labels based, for instance, on Gadolinium(III) or trityl, display either very broad spectra and low modulation depths or very narrow spectra that are unsuitable for the kinds of experiments that we want to conduct.
Malte Drescher, Professor for Spectroscopy of Complex Systems, Department of Chemistry, University of Konstanz
A new study published by Drescher, Wittmann and their team of chemists from the University of Konstanz, which was published online on August 14th in the journal ChemBioChem Communications, presents a new method for labeling proteins that features nitroxide-based spin labels and genetically encoded noncanonical amino acids (ncAAs) as targets for SDSL.
“Nitroxides provide ideal spectral width and access to dynamic information,” says Anandi Kugele, a doctoral researcher at the Konstanz Research School Chemical Biology (KoRS-CB) and first author on the study, who received an esteemed travel grant from the National High Magnetic Field Laboratory to present the findings at the 2019 Rocky Mountain Conference on Magnetic Resonance in Denver, Colorado (USA).
Kugele added, “Traditional nitroxide-based labels have limited redox stability, which is a drawback for in-cell applications. The challenge for us was to increase nitroxide stability and thus to adapt nitroxide-based spin labels for future routine in vivo use.”
For that reason, the scientists created a new spin label that can be attached to proteins by utilizing inverse-electron-demand Diels-Alder (DAinv) cycloaddition to genetically encoded ncAAs, a technique that has been established as suitable to a wide range of in vivo and in vitro applications. To realize nitroxide stability, the scientists additionally used a protection strategy based on photoremovable protecting groups, which are known to safeguard nitroxides and to discharge them when necessary.
The new spin label — called photoactivatable nitroxide for DAinv reaction, or PaNDA for short — is water-soluble, EPR-active, and deprotection-efficient as both in lysate and in vitro as revealed by tests with the two model proteins green fluorescent protein (GFP) and Escherichia coli oxidoreductase thioredoxin (TRX), which is present in almost all identified organisms.
We do need to improve on the method used to deliver the PaNDA spin label to cells and to test labelling and deprotection efficiencies inside the cell, amongst other things. But our research clearly demonstrates that, in principle, the PaNDA label can be used for EPR measurements in challenging biological environments, including the inside of cells.
Malte Drescher, Professor for Spectroscopy of Complex Systems, Department of Chemistry, University of Konstanz
Drescher continued, “Our tests with E. coli lysate are very promising in this respect. This will open up a whole new range of opportunities for the study of proteins by means of EPR spectroscopy.”