Jan 11 2019
An important structural feature of RNA and DNA embedded in water is represented by pairs of positive magnesium ions and negatively charged phosphate groups.
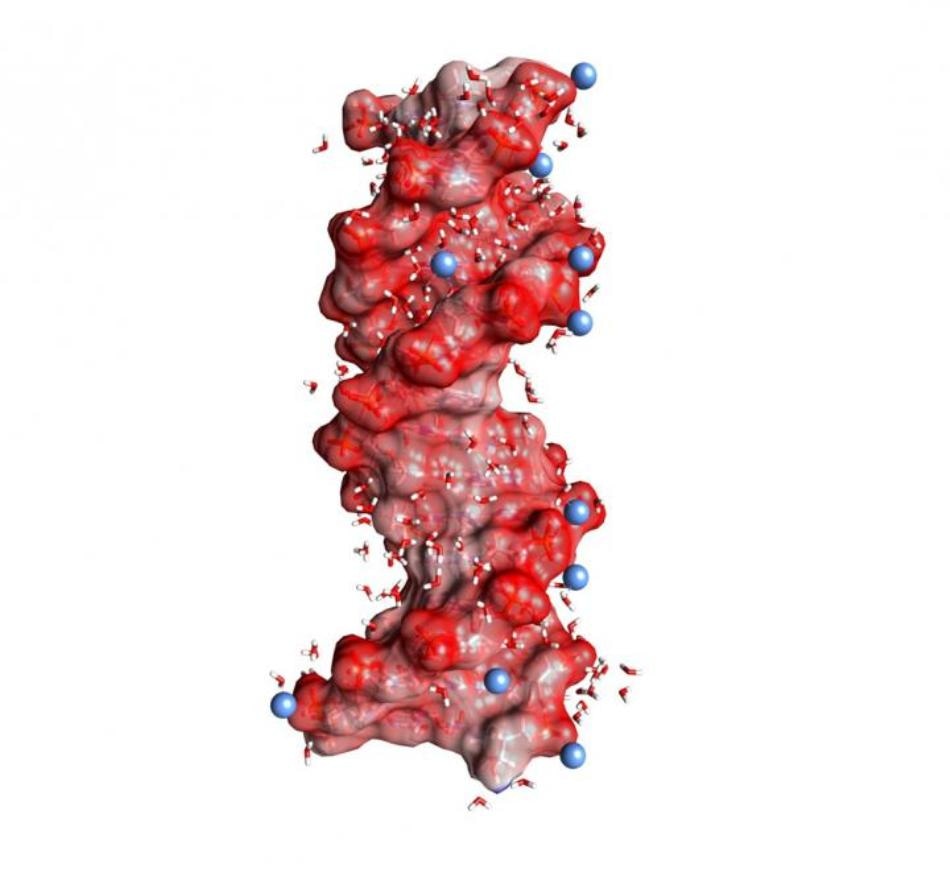 DNA double helix embedded in water (angled small molecules, not to scale). The dark red spheres on the helix surface represent oxygen atoms of the negatively charged PO2- units, the blue spheres positively charged ions in the environment. (Image credit: MBI Berlin)
DNA double helix embedded in water (angled small molecules, not to scale). The dark red spheres on the helix surface represent oxygen atoms of the negatively charged PO2- units, the blue spheres positively charged ions in the environment. (Image credit: MBI Berlin)
Phosphate group vibrations have now been recognized as selective probes for these contact pairs, and they enable mapping the structure and interactions on the ultrafast time scales of molecular dynamics.
The charged polymers—RNA and DNA—serve as key players in protein biosynthesis and encode genetic information in a double helix structure. The negative charges of RNA and DNA are situated in the molecular backbone, which contains ionic phosphate (PO2-) as well as sugar groups. For the macromolecular structures of RNA and DNA to remain stable, the intense repulsive electric forces between the correspondingly charged phosphate groups have to be compensated by ions of opposite, that is, positive charge. In this regard, magnesium (Mg2+) ions are especially relevant because they stabilize the structure, mediate the detection of external binding partners, and also serve as catalytic centers. In addition, macromolecular structural changes through dynamic folding processes are linked to a rearrangement of positive ions integrated into the surrounding water shell.
Positive ions are organized in varied geometries around RNA and DNA: in what is known as site-bound or contact-pair geometries, the location of a positive ion is such that it comes in direct contact with a single oxygen atom of a phosphate group. On the other hand, the so-called outer ion atmosphere contains positive ions that are isolated by at least a single layer of water molecules from the phosphate groups. The functional role of the varied geometries and also the underlying interactions are difficult to understand. For a detailed understanding at the molecular level, highly sensitive probes are required, which help in distinguishing the varied ion geometries without affecting them and also assist in mapping their dynamics on the ultrafast time scale of molecular motions.
Scientists from the Max Born Institute (MBI) have demonstrated in a recent publication that phosphate group vibrations represent noninvasive and sensitive probes of ion geometries in a water setting. Dimethylphosphate (DMP, (CH3O)2PO2-)—a proven model system for the RNA and DNA backbone—was prepared in liquid water with a surplus amount of Mg2+ ions and analyzed using nonlinear vibrational spectroscopy in the femtosecond time domain (1 fs = 10 to the power of -15 s). The experiments utilize two-dimensional infrared (2D-IR) spectroscopy, the most advanced technique for studying the structures and ionic interactions on the intrinsic time scale of varying molecular motions.
Mg2+ ions in direct contact with a PO2- group are mapped by the experiments through a unique feature in the 2D-IR spectrum. The interaction with the Mg2+ ion causes the asymmetric PO2- stretching vibration to shift to a frequency which is greater than the frequency in the absence of Mg2+ ions. The time evolution and lineshape of this novel feature demonstrate the fluctuations of the embedding water shell and the contact ion pair geometry on a time scale of hundreds of femtoseconds, whereas the contact pair itself is prevalent for relatively longer times (~10 to the power of -6 s). A detailed hypothetical analysis demonstrates that the delicate balance of attractive electrostatic (Coulomb) forces and repulsive forces owing to the quantum-mechanical exchange interaction control the frequency position of the phosphate vibration.
The potential of the 2D-IR spectroscopy to define the short-ranged phosphate-ion interaction in solution offers a new analytical tool that complements the structural methods presently available. An extension of this latest technique to RNA and DNA as well as their ionic environment is most promising and anticipated to offer a new understanding about the forces driving folding processes and stabilizing equilibrium structures.