Oct 26 2012
At the edge of the Muir College parking lot sits a structure shaped like an enormous tan igloo encircled by a chain-link fence. An upright wooden post carved into a face like a totem with a sweet, sleepy expression guards the entrance, offering no hint of what goes on within.
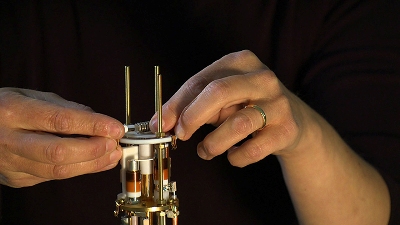 A tiny vial holding a specially prepared sample of protein inserts into a custom-made instrument that allows structural chemists to measure its response to pulses of radio frequencies. Photo UCSD-TV
A tiny vial holding a specially prepared sample of protein inserts into a custom-made instrument that allows structural chemists to measure its response to pulses of radio frequencies. Photo UCSD-TV
Inside, the arching, silvery fabric walls puff between baffles like a down coat. The building is inflatable, with walls filled with air, like a giant Moon Bounce, minus the squishy floor. Inside the open interior sits a series of shiny tanks, shaped like big Thermos bottles, some surrounded by red chain barriers. Signs warn of hazards: no metal tools, no pacemakers.
“This is a very specialized facility,” says Stanley Opella, professor of chemistry and biochemistry and director of the Center for NMR Spectroscopy and Imaging of Proteins, the organization that operates this building.
The purpose of the facility is to create high-resolution images of proteins that represent their shape and arrangement, atom-by-atom. It’s critical information, Opellla says, with a direct link to human health.
“I would say that disease is a problem of proteins,” he says. “Half of all drugs target receptors on the surfaces of cells.” Precise knowledge of the shapes of these receptors will allow drugmakers to tailor small molecules to better fit specific targets, avoiding collateral hits that can cause detrimental side effects.
When chemists want to know the structure of a molecule, they’ll usually purify it and pack it into a uniform crystal, then focus X-rays on the sample. From patterns formed by the diffracted beam, they can identify shapes such as helices.
It’s relatively easy to make crystals of small molecules, and possible for larger ones, like DNA. But proteins are enormous, hundreds of atoms forming a chain of amino acids that twists, bends and folds into complex shapes critical for their functions. Pockets and loops provide surfaces by which they interact with other molecules, and helices spiral through the double layer of phospholipids that make up the membranes of cells.
It’s these proteins, the ones that span cell membranes, that interest Opella the most. They play an important physiological role as intermediaries between signals circulating outside the cell, such as adrenaline or interleukins, which indicate danger or damage, and the molecular machinery within that drives responses such as quickening heart rate or the mobilization of immune responses.
Because proteins flex and bend in response to their environment, Opella wants to see them in their native state, embedded in a membrane. And proteins won’t crystalize that way.
So, he has developed a different approach, in which those Thermos-like tanks play a critical role. Each houses a superconducting magnet, chilled with liquid helium to temperatures approaching absolute zero. Vials of purified protein, embedded in double-layer membranes just as they would be in cells, fit into custom-designed instruments inserted into the core of the magnet.
Within the strong magnetic field, radio frequency pulses perturb the atoms that make up the protein. How each atom responds depends on their proximity to adjacent atoms.
Using this information, Opella’s team pieces together the protein’s shape, atom-by-atom, like a puzzle.
They’re working on several projects at once. One seeks to solve the shape of a bacterial protein that grabs mercury and holds on tightly as it pulls the toxic metal safely into the cell where it can be transformed into a less harmful form.
Another quest is to better understand proteins involved in human health or disease. This week, they announced a major success. For the first time, they have determined the complete structure of an unmodified human G-protein coupled receptor in its native state: embedded in membrane.
The team mapped the arrangement of atoms in a protein called CXCR1, which detects the inflammatory signal interleukin 8 and, through another protein located inside the cell, triggers a cascade of events that can mobilize immune cells, for example.
Their strategy has revealed a new view of this class of receptors. Previous reports have all noted seven helices weaving through the membrane. Opella’s group sees an eighth lying on the membrane surface, a trait that at least some other receptors in this class must share.
And the loops inside and outside of the cell are holding still. “For years, people thought the loops were mobile. They’re not,” Opella said. “The signals we get from the loops aren’t any weaker than the other parts of the protein as they would be if they were waving about.”
CXCR1 has been implicated in the progression of several types of cancer. In one example, preclinical studies have shown that blocking this receptor inhibits undifferentiated stem cells within breast cancer tumors, leading to the death of all tumor cell types and stopping them from seeding new tumors.
Opella and colleagues hope this finding along with continuing studies of changes in this receptor’s configuration as it binds to interleukin 8 and drug candidates will lead to more effective and less harmful cancer treatments.
“I spent 40 years developing the technology to look at a protein in its lipid bilayer,” Opella said. “I accomplished my goal.”