Apr 1 2019
Red blood cells, also known as RBCs, are considered as amazing particles. These cells collect oxygen from the humans’ lungs and carry it throughout their body to sustain them.
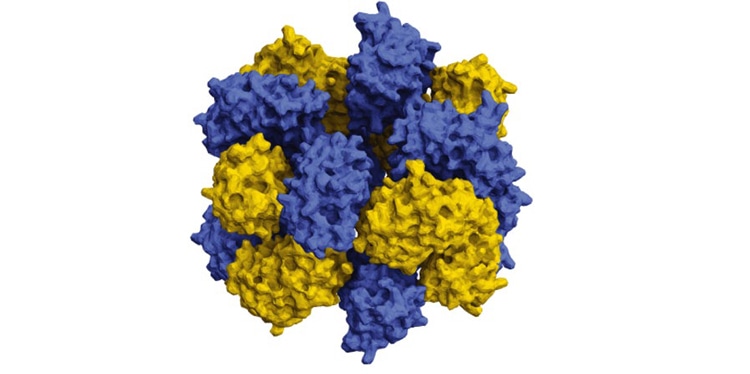 Using supercomputers, scientists are just starting to design proteins that self-assemble to combine and resemble life-giving molecules like hemoglobin. (Image credit: Taylor et al.)
Using supercomputers, scientists are just starting to design proteins that self-assemble to combine and resemble life-giving molecules like hemoglobin. (Image credit: Taylor et al.)
Red blood cells contain the hemoglobin molecule that transports oxygen by altering its shape in an all-or-nothing manner. Within the hemoglobin, four copies of the same protein close and open similar to flower petals, structurally paired to react to one another.
Now, with the help of supercomputers, researchers are just beginning to engineer proteins that have the potential to self-assemble to integrate and look like life-giving molecules, such as hemoglobin. According to the researchers, their techniques can possibly be used in valuable technologies like building materials, pharmaceutical targeting, “smart” sensing, artificial energy harvesting, and so on.
This work was performed by a team of scientists who supercharged the proteins, that is, they changed amino acids—the subunits of proteins—to impart an artificially high negative or positive charge to the proteins. The science team used proteins extracted from jellyfish and successfully assembled an intricate 16 protein structure made up of a pair of stacked octamers by simply supercharging. The results of the study have been reported in the journal Nature Chemistry in January 2019.
In order to validate and inform these experimental outcomes, the team subsequently used supercomputer simulations. Supercomputer allocations on Comet at the San Diego Supercomputer Center (SDSC) and Stampede2 at the Texas Advanced Computing Center (TACC) were granted to the scientists via XSEDE—the Extreme Science and Engineering Discovery Environment supported by the National Science Foundation (NSF).
We found that by taking proteins that don't normally interact with each other, we can make copies that are either highly positively or highly negatively charged. Combining the highly positively and negatively charged copies, we can make the proteins assemble into very specific structured assemblies.
Anna Simon, Study Co-Author and Postdoctoral Researcher, Ellington Lab, UT Austin.
The researchers have dubbed their new strategy “supercharged protein assembly” in which they integrate fabricated supercharged variants to drive defined interactions between proteins.
We exploited a very well-known and basic principle from nature, that opposite charges attract”. Anna Simon’s group found that when they mix these charged variants of green fluorescent protein, they get highly ordered structures. That was a real surprise.
Jens Glaser, Study Co-Author and Assistant Research Scientist, Department of Chemical Engineering, University of Michigan.
Glaser works with the Glotzer Group.
Resembling a braided ring, the stacked octamer structure is made up of 16 proteins—two intertwined rings of eight interacting in highly specific, discrete patches.
“The reason why it’s so hard to engineer proteins that interact synthetically is that making these interacting patches and having them all line up right such that they’ll allow the proteins to assemble into bigger, regular structures is really hard,” Simon explained. The researchers resolved the issue by adding several negative and positive charges to fabricate variants of the green fluorescent protein, or GFP, a well-researched “lab mouse” protein extracted from the Aequorea victoria jellyfish.
In this regard, the positively charged protein, called cerulean fluorescent protein (Ceru) +32 by the scientists, had more prospects to interact with GFP -17—the negatively charged protein.
By giving these proteins all these opportunities, these different places where they could potentially interact, they were able to choose the right ones. There were certain patterns and interactions that were there, available, and energetically favored, that we didn't necessarily predict beforehand that would allow them to assemble into these specific shapes.
Anna Simon, Study Co-Author and Postdoctoral Researcher, Ellington Lab, UT Austin.
In order to obtain the fabricated charged fluorescent proteins, Simon and co-authors Barrett Morrow, Jimmy Gollihar, and Arti Pothukuchy encoded their genes, which also included a chemical tag utilized for purification on portable DNA pieces known as plasmids in E. coli, and subsequently harvested the tagged protein grown by E. coli. After combining the proteins together, the scientists initially believed that the proteins may simply interact to create huge and haphazardly structured clumps.
“But then, what we kept on seeing was this weird, funny peak around 12 nanometers, that was a lot smaller than a big clump of protein, but significantly bigger than the single protein,” stated Simon.
Using a Zetasizer instrument at the Texas Materials Institute of UT Austin, the researchers determined the size of the particles that formed and eventually confirmed that both GFP and cerulean proteins are present in the particles.
Förster Resonance Energy Transfer, or FRET, which is capable of measuring the energy transfer between fluorescent proteins of different colors, generate fluorescence in response to different energies of light to observe if they are proximal together. Negative stain electron microscopy, performed by the team of David Taylor, assistant professor of molecular biosciences at UT Austin, detected the particular structure of the particles. This technique revealed that the 12-nm particle containing a stacked octamer is made up of 16 proteins.
“We found that they were these beautifully shaped flower-like structures,” stated Simon.
With the help of cryo-electron microscopy, Yi Zhou, co-author from Taylor’s team of UT Austin, further increased the resolution to expose the details of the stacked octamer at the atomic level.
The measurements were refined by computational modeling, indicating the way proteins were organized into a vivid picture of the attractive, flower-like structure, Jens Glaser said.
We had to come up with a model that was complex enough to describe the physics of the charged green fluorescent proteins and present all the relevant atomistic details, yet was efficient enough to allow us to simulate this on a realistic timescale. With a fully atomistic model, it would have taken us over a year to get a single simulation out of the computer, however fast the computer was.
Jens Glaser, Study Co-Author and Assistant Research Scientist, Department of Chemical Engineering, University of Michigan.
In order to simplify the model, the researchers decreased the resolution without affecting the critical aspects of protein interactions.
“That’s why we used a model where the shape of the protein is exactly represented by a molecular surface, just like the one that’s measured from the crystallographic structure of the protein,” added Glaser.
What really helped us turn this around and improve what we were able to get out of our simulations was the cryo-EM data. That’s what really helped us find the optimal configuration to put into these simulations, which then helped us validate the stability arguments that we were making, and hopefully going forward make predictions about ways that we can destabilize or modify this structure.
Vyas Ramasubramani, Graduate Student, Department of Chemical Engineering, University of Michigan.
The researchers needed plenty of computing power to perform the calculations on their preferred scale.
We used XSEDE to basically take these huge systems, where you have lots of different pieces interacting with each other, and calculate all of this at once so that when you start moving your system forward through some semblance of time, you could get an idea for how it was going to evolve on somewhat real timescales. If you tried to do the same kind of simulation that we did on a laptop, it would have taken months if not years to really approach understanding whether or not some sort of structure would be stable. For us, not being able to use XSEDE, where you could use essentially 48 cores, 48 compute units all at once to make these calculations highly parallel, we would have been doing this much slower.
Vyas Ramasubramani, Graduate Student, Department of Chemical Engineering, University of Michigan.
At the TACC, the Stampede2 supercomputer contains 4,200 Intel Knights Landing as well as 1,736 Intel Skylake X compute nodes. In each Skylake node, there are 48 cores—the rudimentary unit of a computer processor.
“The Skylake nodes of the Stampede2 supercomputer were instrumental in achieving the performance that was necessary to compute these electrostatic interactions that act between the oppositely-charged proteins in an efficient manner,” stated Glaser. “The availability of the Stampede2 supercomputer was at just the right point in time for us to perform these simulations.”
The scientists initially tested their simulations on the Comet system at the SDSC.
“When we were first figuring out what kind of model to use and whether this simplified model would give us reasonable results, Comet was a great place to try these simulations,” stated Ramasubramani. “Comet was a great testbed for what we were doing.”
Looking at the larger scientific picture, the team believes that this kind of study provides a deeper insight into why a large number of natural proteins will oligomerize, or combine together to create more fascinating and complex structures.
“We showed that there doesn’t need to be a very specific, pre-distinguished set of plans and interactions for these structures to form,” stated Simon. “This is important because it means that maybe, and quite likely we can take other sets of molecules that we want to make oligomerize and generate both positively charged and negatively charged variants, combine them, and have specifically ordered structures for them.”
Bone, shells, and feathers are examples of natural biomaterials that can be strong yet lightweight.
We think supercharged protein assembly is an easier way to develop the kind of materials that have exciting synthetic properties without having to spend so much time or having to know exactly how they’re going to come together beforehand. We think that will accelerate the ability to engineer synthetic materials and for discovery and exploration of these nanostructured protein materials.
Anna Simon, Study Co-Author and Postdoctoral Researcher, Ellington Lab, UT Austin.
The study titled, “Supercharging Enables Organized Assembly of Synthetic Biomolecules,” was published in the journal Nature Chemistry in January 2019. The co-authors of the study are Anna J. Simon, Yi Zhou, Arti Pothukuchy, Jimmy Gollihar, Jillian C. Gerberich, Janelle C. Leggere, Barrett R. Morrow, Cheulhee Jung, David W. Taylor, and Andrew D. Ellington of UT Austin; and Vyas Ramasubramani, Jens Glaser, and Sharon C. Glotzer of the University of Michigan.
The research was funded by the US Army Research Laboratory, US Army Research Office, the Welch Foundation, the Cancer Prevention and Research Institute of Texas, and an Arnold O. Beckman postdoctoral fellowship held by AJS. Sharon Glotzer, the Anthony C. Lembke Department Chair of Chemical Engineering at the University of Michigan; and Andy Ellington, associate director of UT Austin’s Center for Systems and Synthetic Biology are the grant principal investigators. The National Science Foundation funded the XSEDE resources.